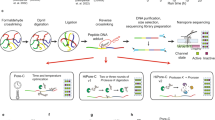
Abstract
The genomes of multicellular organisms are extensively folded into 3D chromosome territories within the nucleus1. Advanced 3D genome-mapping methods that combine proximity ligation and high-throughput sequencing (such as chromosome conformation capture, Hi-C)2, and chromatin immunoprecipitation techniques (such as chromatin interaction analysis by paired-end tag sequencing, ChIA-PET)3, have revealed topologically associating domains4 with frequent chromatin contacts, and have identified chromatin loops mediated by specific protein factors for insulation and regulation of transcription5,6,7. However, these methods rely on pairwise proximity ligation and reflect population-level views, and thus cannot reveal the detailed nature of chromatin interactions. Although single-cell Hi-C8 potentially overcomes this issue, this method may be limited by the sparsity of data that is inherent to current single-cell assays. Recent advances in microfluidics have opened opportunities for droplet-based genomic analysis9 but this approach has not yet been adapted for chromatin interaction analysis. Here we describe a strategy for multiplex chromatin-interaction analysis via droplet-based and barcode-linked sequencing, which we name ChIA-Drop. We demonstrate the robustness of ChIA-Drop in capturing complex chromatin interactions with single-molecule precision, which has not been possible using methods based on population-level pairwise contacts. By applying ChIA-Drop to Drosophila cells, we show that chromatin topological structures predominantly consist of multiplex chromatin interactions with high heterogeneity; ChIA-Drop also reveals promoter-centred multivalent interactions, which provide topological insights into transcription.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The ChIA-Drop datasets, RNAPII ChIA-Drop datasets and RNAPII ChIA-PET datasets have been deposited in the Gene Expression Omnibus database with accession number GSE109355. A manuscript is in preparation on the ChIA-DropBox computational pipeline, and all software will be released as open-source code with that manuscript. All other relevant data are available from the corresponding author upon reasonable request.
References
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Tang, Z. et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627 (2015).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588 (2017).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Zheng, G. X. et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34, 303–311 (2016).
Kharchenko, P. V. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011).
Lajoie, B. R., Dekker, J. & Kaplan, N. The hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75 (2015).
Cover, T. M. & Thomas, J. A. Elements of Information Theory 2nd edn (John Wiley, New York, 2006).
Ulianov, S. V. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26, 70–84 (2016).
Lanzuolo, C., Roure, V., Dekker, J., Bantignies, F. & Orlando, V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9, 1167–1174 (2007).
Szabo, Q. et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv. 4, eaar8082 (2018).
Rowley, M. J. et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852 (2017).
Yang, J., Ramos, E. & Corces, V. G. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 22, 2199–2207 (2012).
van Bemmel, J. G. et al. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 5, e15013 (2010).
Cai, W. et al. RNA polymerase II-mediated transcription at active loci does not require histone H3S10 phosphorylation in Drosophila. Development 135, 2917–2925 (2008).
Duff, M. O. et al. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 521, 376–379 (2015).
Alberts, B. et al. Molecular Biology of the Cell 4th edn (Garland Science, New York, 2002).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018).
Cisse, I. I. et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667 (2013).
Zhao, Z. W. et al. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc. Natl Acad. Sci. USA 111, 681–686 (2014).
Cook, P. R. A model for all genomes: the role of transcription factories. J. Mol. Biol. 395, 1–10 (2010).
Acknowledgements
This study is supported by a Jackson Laboratory Director’s Innovation Fund (DIF19000-18-02). Y.R. and C.-L.W. are funded by 4DN (U54 DK107967) and ENCODE (UM1 HG009409) consortia. Y.R. is also funded by Human Frontier Science Program (RGP0039/2017), and supported by the Florine Roux Endowment.
Reviewer information
Nature thanks David Gifford and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.Z., Y.R. and C.-L.W. conceptualized the ChIA-Drop strategy and designed the studies. M.Z. conducted experiments with assistance from Z.L., R.M., L.G. and C.Y.N. for ChIA-Drop library construction and sequencing. P.W., E.P. and J.J.Z. contributed RNAPII ChIA-PET data. X.R., D.C. and S.Z.T. contributed PacBio results. S.Z.T., M.K., D.C. and B.L. developed the computational pipeline ChIA-DropBox with the assistance of C.H.W. M.Z. and Y.R. wrote the manuscript with input from D.C., M.K., S.Z.T. and B.L. All co-authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 ChIA-Drop method optimization.
The efficiency of the microfluidics system for chromatin DNA barcoding and amplification was characterized by MiSeq sequencing data, in which each test generated 2–4 million sequencing reads. The numbers of captured GEM barcodes, the percentages of uniquely mapped reads, and the read-length distribution are presented for data quality assessment. a, Pure DNA versus chromatin DNA. Both pure DNA and chromatin DNA templates were prepared from the same chromatin sample. The chromatin sample was prepared by in situ HindIII digestion, followed by sonication for nuclear lysis. The chromatin DNA used for test was in a crosslinked state, and some DNA positions were bound by a protein component. The pure DNA was purified from the chromatin fragment after de-crosslinking. The length of the DNA templates was about 3,000 bp. Most of the pure DNA sequencing reads were of maximum length (130 bp); of these reads, 96% were mappable. The chromatin DNA yield 59% mappable reads. b, Distance density comparison of pure DNA and chromatin DNA. The relative probability densities of the log10 of fragment-to-fragment distances in a GEM are plotted, categorized by the number of fragments per GEM (F#), colour-coded from blue to red for fragment numbers F = 2 to F = 11. Pure DNA (left) and chromatin DNA (right) data are both plotted on the same colour scale. The GEMs containing a low number of fragments showed distributions similar to those of the pure DNA, whereas the chromatin DNA with GEMs contain a high number of fragments displayed different patterns. c, Two-dimensional heat map comparison of pure DNA (HindIII, 6-bp cutter) and chromatin DNA (Mbol, 4-bp cutter or HindIII). The pure DNA data show random interactions and a lack of chromatin topological structures; the Mbol chromatin DNA data show some evidence for these structures, and HindIII chromatin DNA data show rich data for abundant chromatin contact structures. d, Chromatin fragment length by different fragmentation methods. Chromatin sample digested by a 4-bp cutter (MboI, ~ 300 bp) or a 6-bp cutter (HindIII, ~3,000 bp), or sheared by sonication (~ 6,000 bp) were prepared accordingly. The longer chromatin fragments (3,000–6,000 bp) generated a larger number of mappable DNA sequencing reads (≥50 bp) than did the shorter fragments. e, Summary statistics of GEMs from chromatin libraries prepared by Mbol and HindIII digestion. The read statistics between the two libraries are comparable under the same loading amount, but the fragment histograms of GEMs are different between the two libraries. The HindIII data generated a larger number of uniquely mappable reads, and of GEMs containing a high number of fragments than did the Mbol data, which contributes to differences seen between the chromatin structures shown in c. f, Chromatin sample loading by different input quantity. An input of 0.5 ng of chromatin DNA yields optimal results. When input was too low (0.5 pg), the majority of the sequencing reads were only 19–20 bp (barcode primer sequence) in length, which indicates that most droplets lack chromatin materials. g, Inter-species chromatin experiment. Chromatin samples of Drosophila S2 and human GM12878 cells were mixed in an equal number of cells or in an equal quantity of chromatin DNA. Barcoded sequencing reads were mapped to each reference genome. Reads with the same GEM barcode were grouped as a GEM. GEMs with fly-only, human-only or mixed reads were identified. The ratio of mixed GEMs to total GEMs provides an approximate likelihood of a mixed chromatin complex in a droplet. When tested with equally mixed numbers of cells, the number of GEMs with chromatin fragments of human origin is 20-fold more than the number of GEMs of Drosophila origin (181,956/9,149 = 19.89), which approximately reflects the ratio of the genome lengths for humans and Drosophila (3,000 Mb (human)/175 Mb (Drosophila) = 17.14). Notably, in the test with equal chromatin mass, the GEMs with mixed origins of fragments were only 5.1% of the total, which indicates that the proportion of droplets with mixed chromatin samples is small.
Extended Data Fig. 2 Reproducibility and characterization of ChIA-Drop data.
a, Genome-wide reproducibility assessment of ChIA-Drop and RNAPII ChIA-Drop. To assess data quality and reproducibility, we conducted a comprehensive comparison between four datasets (ChIA-Drop, Hi-C, RNAPII ChIA-Drop and RNAPII ChIA-PET), each with two replicates for reproducibility analysis, using the program HiCRep. Rows and columns represent the eight datasets of interest, and each cell entry indicates the correlation coefficient (‘scc’ output from HiCRep) between the corresponding row and column. The entries are colour-coded by their values; darker colours represent higher values. b, Scatter plot of insulation scores between ChIA-Drop replicates. The insulation scores of every 10-kb bin (n = 11,918) are computed and plotted genome-wide for both replicates of ChIA-Drop, with Spearman correlation coefficient ρ = 0.83. c, Venn diagram of TAD comparison between ChIA-Drop replicate 1 and replicate 2. Of 471 and 522 TADs called from replicate 1 and replicate 2, respectively, 79% overlapped. Box plots of insulation scores for reproducible TADs and replicate-specific TADs show that the reproducible TADs have higher scores. Box plot, middle line denotes median; box denotes IQR; and whiskers denote 1.5× IQR. For both of the replicates, the unique and intersecting parts of the Venn diagram are highly different from each other. Two-sided Wilcoxon test P value < 2.2 × 10−16 for replicate 1; P value = 5.42 × 10−13 for replicate 2. Further details are provided in the Supplementary Methods. d, Browser view of A and B compartment, and TAD of ChIA-Drop and Hi-C data. For a given 1.2-Mb window, ChIA-Drop and Hi-C data heat maps are presented along with their TADs (triangle), and A (light orange) and B (grey) compartments, by eigenvectors, and with histone ChIP–seq H3K27ac (golden) and H3K27me3 (black) tracks. ChIA-Drop and Hi-C data exhibit similar patterns to one another. e, Epigenomic patterns in A and B compartments. Each of the A and B compartments is binned into 300 bins, and both compartments are extended by 100 bins on both ends. The densities of H3K27ac and H3K27me3 ChIP–seq signals, scaled by reads per million (RPM), are plotted. f, TAD size distribution. TADs were called via insulation scores from Hi-C and two replicates of ChIA-Drop data. The resulting densities of TAD sizes (lengths) are plotted, with a peak at 142 kb for all three datasets. g, Two-dimensional contact map of two replicates of ChIA-Drop data. A 10-Mb region is shown for two replicates in contact heat maps. HiCRep correlation coefficient (scc) = 0.79. h, Distribution of contact distances by fragment size. The empirical cumulative distribution function of the log10 of contact distances supports the observed pattern that small complexes have larger contact distances than large complexes. Colour bar scales from blue to red for the number of fragments, in ascending order. F#, number of fragments per complex. i, Distribution plots for the number of fragments per chromatin complex involved in TADs or in gap regions. j, A magnified browser view (1.5 Mb) showing that ChIA-Drop complex clusters associate with TADs. From top to bottom, ChIA-Drop 2D pairwise contact map showing a region with a number of TADs and boundary regions; A and B (orange and grey, respectively) compartments called by eigenvectors from ChIA-Drop data; cluster view (brown colour) of the chromatin complexes in this region with binning at 10-kb resolution; and a fragment view of the chromatin complexes in this region. Each set of the assorted colour bars connected by a straight line depicts a ChIA-Drop chromatin complex. A further magnified fragment view (490 kb), highlighting the inter-TAD chromatin complexes, is shown in a separate panel at the bottom.
Extended Data Fig. 3 Technical validation of ChIA-Drop data.
a, Comparison of ChIA-Drop and PacBio detected multiple chromatin complexes. In a given 270-kb window, a linear fragment view of ChIA-Drop data shows clusters associated with TAD structures. A magnified view displays the overlapping chromatin fragments from ChIA-Drop and PacBio complexes, with matching regions highlighted in light blue. Data for complexes from both methods exhibit a high level of heterogeneity. b, Comparison of the distributions of fragment number in chromatin complexes in PacBio and ChIA-Drop data. Both methods captured multiplex chromatin complexes at a single-molecule resolution. For the same cost, PacBio sequencing generated a smaller number of reads than ChIA-Drop sequencing using MiSeq. With more reads, the ChIA-Drop data show a higher multiplexity in chromatin contacts than revealed in the PacBio data. c, ChIA-Drop data of chromatin contacts with multiple fragments are shown in a pairwise 2D contact map (top), as complex clusters (middle) and as fragment views (bottom) in this 560-kb region in chromosome X. The same region has previously been studied15 using 3D super-resolution DNA-FISH, with three probes (1, green; 2, red; 3, blue) designed to test intra-TAD contacts (probe 1–probe 2) and inter-TAD contacts (probe 1–probe 3, probe 2–probe 3 and probe1–probe 2–probe 3). ChIA-Drop detected all possible pairwise combinations and three-way contacts, and the number of GEMs are noted as n values. d, ChIA-Drop contact frequencies between the three loci are plotted. The highest contacts were between the intra-TAD loci 1 and 2, and the three other combinations of inter-TAD were low (approximately at the same level). The intra-TAD and inter-TAD contact frequencies matched with the physical distances of the three loci as measured by 3D super-resolution DNA-FISH. More specifically, individual GEMs contained fragments that overlap the three probed loci were detected, which validates ChIA-Drop for detecting multiplex chromatin contacts. e, Diagram of the Hox gene cluster BX-C (bithorax complex, which comprises three Hox genes, Ubx, abd-A and Abd-B). The pairwise chromatin contacts between Fab-7 and Abd-B, and Fab-7 and abd-A, are shown by two-colour FISH in S2 cells, but not in S3 cells14. The three loci, bxd, Fab-7 and the Abd-B promoter (Pm), were used to make fluorescent DNA probes as shown. Fab-7 and bxd are separated by approximately 130 kb, and Fab-7 and Abd-B promoter are separated by approximately 70 kb. Gene Antp in the Hox gene cluster ANT-C (Antennapedia complex), located approximately 10 Mb away from BX-C, was also included, allowing a fluorescent probe to provide a nuclear position point in the FISH experiment. Arrows indicate gene orientation; orange squares indicate promoter regulatory elements; and coloured circles indicate DNA probes of corresponding colours. f, Four-colour FISH in Drosophila S2 cells. The three loci (Abd-B, Fab-7 and bxd; n = 20, measured in 179 nuclei from 2 independent biological replicates) were colocalized, and the probe for Antp locus provided spatial nuclear position. Right panel provides a magnified view of yellow square in the left panel. g, Four-colour FISH in Drosophila S3 cells. The 3 loci (Abd-B, Fab-7 and bxd; n = 1, measured in 76 nuclei from 2 independent biological replicates) were not colocalized, which reveals the topological structure of this Hox gene cluster in S3 cells that is different from the structure in S2 cells as shown in f. As with the S2 cell, the probe for the Antp locus provided spatial nuclear position. h, Curves of spatial distances (in μm) between pairs of probes, measured by Imaris image analysis software v.9.2. The mode of the distance between Abd-B and Fab-7 is 0.24 μm in S2 cells (n = 179), and 0.41 μm in S3 cells (n = 76); the mode of the distance between Fab-7 and bxd is 0.28 μm in S2 cells (n = 179), and 0.3 μm in S3 cells (n = 76). In both cells, the modes of the distance between Fab-7 and Antp are larger than 1.81 μm. i, Histogram displays the ChIA-Drop-identified chromatin contacts at the Hox gene locus between Abd-B to Fab-7, Fab-7 to bxd, and Abd-B to Fab-7 to bxd together simultaneously. j, Histogram displays the proportions of nuclei that detected colocalization at loci between Abd-B and Fab-7, between Fab-7 and bxd, and between Abd-B, Fab-7 and bxd simultaneously. With a spatial-distance cut-off of 0.24 μm, more nuclei were found to be colocalized in S2 cells (n = 179) than in S3 cells (n = 76). k, Distributions of spatial distances (in μm) between pairs of probes, measured using Imaris image analysis software. The modes of the distance distributions for each pair of probes are as follows: T4a–T4b, mode = 0.22 (nuclei, n = 221); T3–T4a, mode = 0.5 (nuclei, n = 221); T3–T4b, mode = 0.5 (nuclei, n = 404); T2*–T4b, mode = 0.78 (nuclei, n = 404); and T1–T4b, mode = 0.63 (nuclei, n = 404). l, Spatial distances (in μm) of probe pairs in normal S2 cells (n = 213) and heat-shock-treated cells (n = 150), analysed using Imaris image analysis software. Top, distributions of spatial distances in normal, untreated cells (n = 213). The modes of the spatial-distance distributions for each of the probe pairs are: R1–R2, mode = 0.48 and R1–R4, mode = 0.81. Bottom, curves of spatial distances in the heat-shock-treated cells (n = 150). The modes of the spatial-distance distributions for each of the probe pairs are: R1–R2, mode = 0.57 and R1–R4, mode = 1.06. The spatial distances of R1–R2 and R1–R4 observed in heat-shock-treated cells were notably larger than those observed in untreated normal cells.
Extended Data Fig. 4 Characterization of RNAPII ChIA-Drop data.
a, Comparison of 2D contact maps between ChIA-Drop and RNAPII ChIA-Drop replicates 1 and 2, in a 1.8-Mb segment of chromosome 3L. TADs called in ChIA-Drop data are shown in 2D contact maps, along with the tracks of histone marks H3K27ac and H3K27me3, and RNA-seq, to illustrate the active and inactive regions. The complex clusters of RNAPII ChIA-Drop data are shown at the bottom. Notably, contact signals in RNAPII ChIA-Drop data were enriched in active regions but reduced in inactive regions. b, Distribution of contact distances by GEM complexity of fragment number. The empirical cumulative distribution function of the log10 of contact distances supports the pattern that small complexes have larger contact distances than large complexes. Colour scales for each curve were from the spectrum of blue to red for fragment numbers 2 to 50, in ascending order. F#, number of fragments per complex. c, Basic browser view in a 37-kb window (chromosome 2R: base pairs 5,423,000 to 5,460,000) to display the loops and peaks from two replicates of RNAPII ChIA-Drop data. Multiple ChIA-Drop fragments (n = 79) of chromatin complexes (n = 26) are shown in the bottom panel. d, Scatter plot shows peak (n = 14,347) correlations between the two replicates, inside RAID regions. Pearson correlation coefficient, r = 0.92. e, Venn diagram shows the loop anchor overlap between the two replicates, inside RAID regions. PET count cut-offs for loop numbers are indicated.
Extended Data Fig. 5 Characterization of RNAPII ChIA-PET data.
a, Correlation of RNAPII ChIA-PET data between two replicates. Scatter plot shows two replicates of RNAPII ChIA-PET data, peak coverage (RPM, top, n = 5,924 union peaks), loop frequency (RPM, middle, n = 23,773 overlapped loops) and 2D contact map (100 kb per bin, RPM, bottom, n = 1,449,616 bins). Pearson correlation coefficients are r = 0.976, r = 0.928 and r = 0.939 for the top, middle and bottom panels, respectively. b, Loop span distribution by PET counts. The interaction loop spans (n = 1,461,514) are recorded separately by PET counts, and their distributions are plotted. Interactions with high PET counts tend to have shorter loop spans than those with low PET counts. Colours denote PET counts. c, Mixture model for loop span. The population of loops (n = 17,088 loops) is separated into mid-range interaction (red) and long-range interaction (green) classes, using the Gaussian mixture model. The dotted line shows the marginal distribution of the log10(loop span) for loops with ≥4 PET counts; 100 kb is selected as a cut-off threshold. d, Relationship between loop size and PET counts with respect to RAIDs. Loops (n = 90,885, PET ≥ 2) are categorized as intra-RAID (n = 476 RAIDs) if both anchors are in the same RAID and as inter-RAID if they span two different RAIDs. The log of loop size and log of PET counts are plotted; colours indicate the two categories. e, Browser view. A 737-kb region is presented along with RAIDs, epigenomically defined states (ChromHMM; colour codes indict nine chromatin states), and various ChIP–seq and RNA-seq signal tracks. Daisy-chain-like loops tend to be clustered inside a RAID, but long-range inter-RAID loops also exist. A 59-kb RAID region is highlighted in the bottom panel. f, Characteristics of RAIDs (n = 476). The lengths, loop PET counts and gene counts in RAIDs are plotted as histograms and density lines.
Extended Data Fig. 6 Comparison of RNAPII ChIA-PET and RNAPII ChIA-Drop.
a, Comparison of 2D contact maps. The 2D contact maps of RNAPII ChIA-PET (lower left of each panel) and RNAPII ChIA-Drop (upper right of each panel) are displayed at multiple resolutions (for the whole chromosome 2R, a 10-Mb segment of chromosome 2R and two 2.5-Mb domains of chromosome 2R). Dotted circles highlight the long-range loops identified by both methods. b, Loops, peaks and fragment visualization. In a 63-kb window in chromosome 2R, the RNAPII ChIA-PET loops and peaks are similar for Kc167 and S2 cell lines. Moreover, the RNAPII ChIA-Drop data in loop and peak views are in high concordance with the RNAPII ChIA-PET loops in S2 cells. Individual complexes with multiplex fragments show further details of RNAPII ChIA-Drop data. C#, complex number; F#, fragment number. c, Peak correlation. The signal density of peak unions (n = 1,521) in RPM is plotted using a log10 scale, taking in account all peaks called by RNAPII ChIA-PET and RNAPII ChIA-Drop data inside RAIDs (n = 476). Pearson correlation coefficient, r = 0.69. d, Loop count correlation. The number of loops in RAIDs (n = 476) identified either in RNAPII ChIA-Drop or RNAPII ChIA-PET data were counted for each RAID. The loop counts of each RAID were plotted. Pearson correlation coefficient, r = 0.85. e, Promoter-to-promoter (PP) correlation. For each gene promoter (n = 9,133 genes with promoter-to-promoter interaction), the number of interactions with another promoter were counted in both RNAPII ChIA-PET and RNAPII ChIA-Drop data, and are plotted using a log10 scale. Pearson correlation coefficient, r = 0.76. f, Promoter-to-enhancer (PE) correlation. Each gene promoter (n = 7,115 genes with promoter-to-enhancer interaction) is evaluated for its interactions with enhancers, and the counts are plotted. Pearson correlation coefficient, r = 0.71. g, Loop anchor correlation. Venn diagram shows that 82.9% of the RNAPII ChIA-PET loops with PET ≥ 3 within RAIDs overlap with 60.5% of the RNAPII ChIA-Drop loops with PET ≥ 4 inside RAIDs. h, Box plot of normalized ChIA-Drop fragment coverage in chromatin domains. Left, coverage in active domains (dark green) and inactive domains (dark grey) from ChIA-Drop data or RNAPII-enriched ChIA-Drop data. The active and inactive domains were called by segmenting signals for RNA-seq and histone modification ChIP–seq (inactive domains, n = 706; active domains, n = 708; ChIA-Drop, two-sided Wilcoxon test P value = 0.72; RNAPII ChIA-Drop, P value = 6.94 × 10−14). i, Box plot of coverage in chromatin domains. The coverage in TADs (grey) and RAIDs (blue) from ChIA-Drop data or RNAPII ChIA-Drop data (TADs, n = 583; RAIDs, n = 476; ChIA-Drop, two-sided Wilcoxon test P value = 0.97; RNAPII ChIA-Drop, P value = 4.61 × 10−5). RAIDs are chromosome regions defined by merging high-confidence loops (PET ≥ 4, span ≤ 100 kb) from RNAPII ChIA-PET data (Supplementary Methods). TADs were called by calculating insulation score. Detailed statistics are provided in the Supplementary Methods. j, Box plot of the complexity of RNAPII ChIA-Drop chromatin complexes in RAIDs. RAIDs (n = 476) are binned at 5-kb resolution and the number of bins covered by each chromatin complex in RNAPII ChIA-Drop data are counted. An expected random distribution based on a binomial model is shown in grey, and green dots are individual data points from ChIA-Drop. Chromatin complexes from RNAPII ChIA-Drop in RAIDs show lower complexity (that is, are more constrained) than expected. k, Box plot of GEM complexity of ChIA-Drop chromatin complexes in TADs. TADs (n = 582) are binned at 5-kb resolution and the number of bins covered by each chromatin complex in ChIA-Drop data are counted. Box plots follow the same convention as in j, but unlike in j, chromatin complexes from ChIA-Drop in TADs show a higher complexity than expected (Supplementary Note 2). l, Bias towards active genes in RNAPII ChIA-Drop data. The number of chromatin complexes (GEM counts) that overlap with active (n = 7,334) and inactive (n = 3,210) gene promoters inside RAIDs are recorded, and normalized by the number of unique isoforms (see Supplementary Methods). The box plot shows a preference towards active genes. Two-sided Wilcoxon test P value <2.2 × 10−16. Detailed statistics are provided in the Supplementary Methods. m, Bias towards active genes in RNAPII ChIA-PET data. Instead of the GEM counts in RNAPII ChIA-Drop (l), the coverage in RNAPII ChIA-PET is recorded and normalized as in l. The box plot for RNAPII ChIA-PET data also skews towards active genes (n = 7,334 active genes and n = 3,210 inactive genes). Two-sided Wilcoxon test P value <2.2 × 10−16. Detailed statistics are provided in the Supplementary Methods. n, Correlation between scRNA-seq and bulk-cell RNA-seq. Using the RPKM values of scRNA-seq and bulk-cell RNA-seq expression levels for genes in S2 cells (n = 2,094 genes), the two sets of numbers are plotted in log10 scale. Pearson correlation coefficient, r = 0.72. o, Visualization of scRNA-seq and bulk-cell RNA-seq. The bulk-cell RNA-seq expression level, and the accumulated scRNA-seq expressed level, have a similar profile to one another. In all box plots, the middle line denotes the median, the box denotes the IQR and the whiskers denote 1.5× IQR. Data points are plotted as individual dots if they are located beyond ends of the whiskers. Further details are provided in the Supplementary Methods.
Extended Data Fig. 7 Examples of RNAPII ChIA-Drop data.
a, Promoter-centric view of RNAPII ChIA-Drop complexes at the 188-kb region, with 28 genes (including lilli, a large gene of 65 kb in length). This region contains 518 promoter-linked complexes, including one-promoter complexes (1P complex, n = 425), two-promoter complexes (2P complexes, n = 89) and three-promoter complexes (3P complexes n = 4). Two-promoter and three-promoter complexes are shown in the bottom two panels. b, Among the chromatin complexes involving three promoters, different subgroups were identified on the basis of expression levels of the three genes. Each chromatin complex is displayed as a dot, arranged by similarity using dimensionality reduction and density-based clustering (Supplementary Methods). Clusters are colour-coded. c, Three-gene complex with one dominant gene. RNAPII ChIA-Drop data connected promoters of three genes: RpL28, CG12078 and ckd. On the basis of RNA-seq and histone marks, only RpL28 displays active behaviours. The expression levels (in RPKM) of the three genes are shown as a bar chart. d, Three-gene complex with three dominant genes. All three genes—CG1115 (also known as Vps37B), Kat60 and CG12713—were simultaneously captured by RNAPII ChIA-Drop data, and are active with similarly high levels of expression. e, Three-gene complex with two dominant genes. Only Aats-asp (also known as AspRS) and NAT1 are active, with Mdr49 potentially having a role as an enhancer (expression level is shown as a bar chart, on the right). f, An example of intra-genic RNAPII ChIA-Drop data. At the lilli locus, processive multiplex chromatin contacts linked promoters that cover the entire gene body are shown. The number of fragments in a complex reflects the potential looping complexity for the one-sided extrusion model in transcription.
Extended Data Fig. 8 Additional examples of RNAPII ChIA-Drop data for one-sided extrusion model in transcription.
a, Browser view of luna gene. Processive multiplex chromatin contacts linked to promoters that cover the entire luna gene body are shown. Only a few chromatin contacts appear upstream of the luna gene, which indicates the specific chromatin interactions in the direction of gene transcription. b, Browser view of shn gene. Similar to the luna gene, the shn locus displays processive multiplex chromatin contacts linked to promoters. Two-dimensional contact maps from RNAPII ChIA-PET data show a stripe pattern of chromatin contact, which supports the RNAPII ChIA-Drop data. c, Processive multiplex chromatin contacts linked to promoters, at the mam locus. d, Processive multiplex chromatin contacts linked to promoters, at the locus of the Nipped-A gene and of the d4 gene, in the opposite direction of transcription.
Supplementary information
Supplementary Information
This PDF file contains Supplementary Methods, Supplementary Notes (1 and 2). Supplementary Methods: I. Experimental Methods, II. Computational Methods and Statistical Analysis. The Supplementary Methods contains the detail of experimental methods, computational methods, and statistical analysis. Supplementary Notes: 1. ChIA-DropBox -- A Data Processing and Visualization Pipeline for ChIA-Drop, 2. Detailed Analysis Methods and Results. The Supplementary Notes contain two parts: one is ChIA-DropBox, the computational pipeline for processing and visualizing ChIA-Drop data; the other part is the detailed analysis methods and results.
Supplementary Table
Supplementary Table 1: Dataset used in this study and ChIA-Drop library information. 1a. Summary of datasets (S2 cell line) used in this study, 1b. Sequencing information of ChIA-Drop. This table contains the information of datasets used in this study and sequencing statistics of ChIA-Drop libraries.
Supplementary Table
Supplementary Table 2: ChIA-Drop complexes (F ≥ 10). This table contains information (genomic span and coordinates) of the chromatin complexes with 10 or more fragments in each complex detected by ChIA-Drop in Drosophila S2 cells.
Supplementary Table
Supplementary Table 3: RNAPII ChIA-Drop complexes (F ≥ 6). This table contains information (genomic span and coordinates) of the chromatin complexes with 6 or more fragments in each complex detected by RNAPII ChIA-Drop in Drosophila S2 cells.
Supplementary Table
Supplementary Table 4: Summary of chromatin domains. 4a. Information of RNAPII associated interaction domains (RAIDs), 4b. Information of Active domains, 4c. Information of Inactive domains. 4d. Information of TADs called from Hi-C, 4e. Information of TADs called from ChIA-Drop (Rep1), 4f. Information of TADs called from ChIA-Drop (Rep2). This table contains (with 6 sub-tables) the genome coordinate information of chromatin domains, including RAIDs by RNAPII ChIA-PET; Active and Inactive domains by histone marks, and TADs called by Hi-C and ChIA-Drop data, respectively.
Supplementary Table
Supplementary Table 5: Genes in 3-promoters multiplex complexes. This table contains a list of genes involved in three-promoter multiplex complexes, and assignment to the 4 gene clustering groups (related to Fig. 4c and Extended Fig. 7b).
Rights and permissions
About this article
Cite this article
Zheng, M., Tian, S.Z., Capurso, D. et al. Multiplex chromatin interactions with single-molecule precision. Nature 566, 558–562 (2019). https://doi.org/10.1038/s41586-019-0949-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0949-1
This article is cited by
-
Single-cell multiplex chromatin and RNA interactions in ageing human brain
Nature (2024)
-
Computational methods for analysing multiscale 3D genome organization
Nature Reviews Genetics (2024)
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
-
High-throughput Pore-C reveals the single-allele topology and cell type-specificity of 3D genome folding
Nature Communications (2023)
-
Evaluating the role of the nuclear microenvironment in gene function by population-based modeling
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.