Abstract
Understanding host interactions that lead to pathogen transmission is fundamental to the prediction and control of epidemics1,2,3,4,5. Although the majority of transmissions often occurs within social groups6,7,8,9, the contribution of connections that bridge groups and species to pathogen dynamics is poorly understood10,11,12. These cryptic connections—which are often indirect or infrequent—provide transmission routes between otherwise disconnected individuals and may have a key role in large-scale outbreaks that span multiple populations or species. Here we quantify the importance of cryptic connections in disease dynamics by simultaneously characterizing social networks and tracing transmission dynamics of surrogate-pathogen epidemics through eight communities of bats. We then compared these data to the invasion of the fungal pathogen that causes white-nose syndrome, a recently emerged disease that is devastating North American bat populations13,14,15. We found that cryptic connections increased links between individuals and between species by an order of magnitude. Individuals were connected, on average, to less than two per cent of the population through direct contact and to only six per cent through shared groups. However, tracing surrogate-pathogen dynamics showed that each individual was connected to nearly fifteen per cent of the population, and revealed widespread transmission between solitarily roosting individuals as well as extensive contacts among species. Connections estimated from surrogate-pathogen epidemics, which include cryptic connections, explained three times as much variation in the transmission of the fungus that causes white-nose syndrome as did connections based on shared groups. These findings show how cryptic connections facilitate the community-wide spread of pathogens and can lead to explosive epidemics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All raw data points are contained within the main-text Figs. and Extended Data Figs. All other data are available from the corresponding author upon reasonable request.
Change history
17 January 2019
In Fig. 3d this Letter, the R2 value should have been ‘0.19’ instead of ‘0.66’; this has been corrected online.
References
Woolhouse, M. E. J. et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc. Natl Acad. Sci. USA 94, 338–342 (1997).
Fraser, C., Riley, S., Anderson, R. M. & Ferguson, N. M. Factors that make an infectious disease outbreak controllable. Proc. Natl Acad. Sci. USA 101, 6146–6151 (2004).
Edmunds, W. J., O’Callaghan, C. J. & Nokes, D. J. Who mixes with whom? A method to determine the contact patterns of adults that may lead to the spread of airborne infections. Proc. R. Soc. Lond. B 264, 949–957 (1997).
Snow, J. On the Mode of Communication of Cholera (John Churchill, London, 1855).
Salathé, M. & Jones, J. H. Dynamics and control of diseases in networks with community structure. PLOS Comput. Biol. 6, e1000736 (2010).
Mossong, J. et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 5, e74 (2008).
Nunn, C. L. & Altizer, S. Infectious Diseases in Primates: Behavior, Ecology, and Evolution (Oxford Univ. Press, Oxford, 2006).
Keeling, M. The implications of network structure for epidemic dynamics. Theor. Popul. Biol. 67, 1–8 (2005).
Altizer, S. et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 34, 517–547 (2003).
Sah, P., Leu, S. T., Cross, P. C., Hudson, P. J. & Bansal, S. Unraveling the disease consequences and mechanisms of modular structure in animal social networks. Proc. Natl Acad. Sci. USA 114, 4165–4170 (2017).
King, A. A., Ionides, E. L., Pascual, M. & Bouma, M. J. Inapparent infections and cholera dynamics. Nature 454, 877–880 (2008).
Lloyd-Smith, J. O. et al. Epidemic dynamics at the human–animal interface. Science 326, 1362–1367 (2009).
Lorch, J. M. et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480, 376–378 (2011).
Langwig, K. E. et al. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol. Lett. 15, 1050–1057 (2012).
Langwig, K. E. et al. Invasion dynamics of white-nose syndrome fungus, midwestern United States, 2012–2014. Emerg. Infect. Dis. 21, 1023–1026 (2015).
Jones, K. E. et al. Global trends in emerging infectious diseases. Nature 451, 990–993 (2008).
Bansal, S., Grenfell, B. T. & Meyers, L. A. When individual behaviour matters: homogeneous and network models in epidemiology. J. R. Soc. Interface 4, 879–891 (2007).
Gurley, E. S. et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 13, 1031–1037 (2007).
Alexander, K. A. et al. What factors might have led to the emergence of Ebola in West Africa? PLoS Negl. Trop. Dis. 9, e0003652 (2015).
Weber, N. et al. Badger social networks correlate with tuberculosis infection. Curr. Biol. 23, R915–R916 (2013).
Wendland, L. D. et al. Social behavior drives the dynamics of respiratory disease in threatened tortoises. Ecology 91, 1257–1262 (2010).
McBryde, E. S. & McElwain, D. L. A mathematical model investigating the impact of an environmental reservoir on the prevalence and control of vancomycin-resistant enterococci. J. Infect. Dis. 193, 1473–1474 (2006).
Poutanen, S. M. et al. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348, 1995–2005 (2003).
Eubank, S. et al. Modelling disease outbreaks in realistic urban social networks. Nature 429, 180–184 (2004).
Lloyd-Smith, J. O. et al. Should we expect population thresholds for wildlife disease? Trends Ecol. Evol. 20, 511–519 (2005).
Cross, P. C. et al. Integrating association data and disease dynamics in a social ungulate: bovine tuberculosis in African buffalo in the Kruger National Park. Ann. Zool. Fenn. 41, 879–892 (2004).
Thomas, D. The physiological ecology of hibernation in vespertilionid bats. Symp. Zool. Soc. Lond. 67, 233–244 (1995).
Langwig, K. E. et al. Drivers of variation in species impacts for a multi-host fungal disease of bats. Phil. Trans. R. Soc. Lond. B 371, 20150456 (2016).
Lloyd-Smith, J. O., Schreiber, S. J., Kopp, P. E. & Getz, W. M. Superspreading and the effect of individual variation on disease emergence. Nature 438, 355–359 (2005).
Edmunds, W. J., Kafatos, G., Wallinga, J. & Mossong, J. R. Mixing patterns and the spread of close-contact infectious diseases. Emerg. Themes Epidemiol. 3, 10 (2006).
Langwig, K. E. et al. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc. R. Soc. Lond. B 282, 20142335 (2014).
Hunt, A., Collins-Poythress, J. & Langwig, K. E. Swabbing protocol for Geomyces destructans https://www.youtube.com/watch?v=KU1EJPJXNPk (2013).
Muller, L. K. et al. Bat white-nose syndrome: a real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans. Mycologia 105, 253–259 (2013).
Fujita, M. S. & Kunz, T. H. Pipistrellus subflavus. Mamm. Species 228, 1–6 (1984).
Dawson, D. L. Functional interpretations of the radiographic anatomy of the femora of Myotis lucifugus, Pipistrellus subflavus, and Blarina brevicauda. Am. J. Anat. 157, 1–15 (1980).
Caceres, M. C. & Barclay, R. M. R. Myotis septentrionalis. Mamm. Species 634, 1–4 (2000).
Kembel, S. W. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010).
Warnecke, L. et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc. Natl Acad. Sci. USA 109, 6999–7003 (2012).
Czenze, Z. J., Jonasson, K. A. & Willis, C. K. R. Thrifty females, frisky males: winter energetics of hibernating bats from a cold climate. Physiol. Biochem. Zool. 90, 502–511 (2017).
Bürkner, P.-C. brms: an R package for bayesian multilevel models using stan. J. Stat. Softw. 80, 1–28 (2017).
Bates, D., Maechler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D. & R Core Team. nlme: linear and nonlinear mixed effects models. R package version 3.1-137 https://cran.r-project.org/web/packages/nlme/index.html (2018).
Acknowledgements
Financial support was provided by the NSF (grants DGE-0741448, DEB-1115895 and EF-0914866), USFWS and Bat Conservation International. E. McMaster provided support and site access; B. Lyon, J. Nicol, L. Childs and E. Palsson provided comments on analyses or the manuscript; and M. Hee, H. Guedi, S. Yamada, K. George, G. Auteri, J. Bailey and N. Tran aided in the laboratory or field.
Reviewer information
Nature thanks B. Fenton, J. Lloyd-Smith and D. Reeder for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.R.H., K.E.L. and A.M.K. conceptualized the project, analysed the data and wrote the paper with input from all authors. J.R.H., K.E.L., J.P.W., H.M.K., J.A.R., A.K., J.E.D., W.H.S., W.F.F. and A.M.K. designed, organized and performed the field study. J.R.H., K.L.P. and J.T.F. performed laboratory testing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Application and re-sighting of UVF dust on bats and in the environment.
a, Applying UVF dust to P. subflavus. b, A dusted P. subflavus shortly after release. c, M. septentrionalis after UVF-dust application. d, A group of eight M. lucifugus roosting together in March. Each bat in the group had UVF dust. e, A dusted M. lucifugus in a group of three bats during March. At least three of the bats in the photo can be seen with UVF dust. f, Location in the environment with patches of UVF dust in March next to a hibernating P. subflavus.
Extended Data Fig. 2 Trends of population counts and timing of UVF-dust and disease data collection.
The grey bar shows the winter period when the UVF-dust study was conducted and the red bar shows the year the P. destructans arrived. Temporal patterns of social group size are provided in Extended Data Fig. 6a.
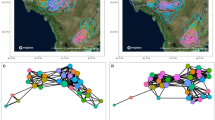
Extended Data Fig. 3 Social and epidemic networks for four additional communities of hibernating bats.
a–h, Each row shows two networks (physical contact and UVF dust) for each of four sites (WI-ST, WI-SB, MI-TM and MI-GA). Each circle (node) represents an individual bat, with colour indicating species; larger numbered circles (1–7) are bats that were dusted with UVF dust with unique colours in November at each site. Lines (edges) between nodes indicate physical contact among bats in social networks (a–d) or UVF dust that originated from the UVF-dusted bat (large circle) in epidemic networks (e–h). Each UVF-dust epidemic network in e–h shows seven replicate epidemics, with each epidemic originating from a single, numbered UVF-dusted bat. Locations of nodes (bats) in the two networks are identical, so the spread of UVF dust within and between groups of touching bats can be visualized in the UVF-dust network.
Extended Data Fig. 4 Comparison of observed data from late-winter and total shared-group connections including group rearrangements.
The y axes show the percentage of individuals of each species at each site observed in direct physical contact, in shared groups or with UVF dust originating from a single focal individual. a, M. lucifugus. b, M. septentrionalis. c, P. subflavus. Small points show the observed (grey) data and augmented (coloured) connections, including group rearrangements. The black open points show the mean late winter estimate of direct contact and social-group connections based on a single time point of the populations without group switching (Fig. 1, Extended Data Fig. 3; left panels). The large coloured points (means in Fig. 2) show the mean number of connections from simulations in which bats rearranged during six arousals over the winter (also shown in Fig. 2). Where the large coloured points are within the black open points, there were no differences between the observed and augmented data including group rearrangements. The connections observed in UVF-dust epidemics are shown for comparison, but do not differ.
Extended Data Fig. 5 Shared-group (cluster) size for four species of hibernating bats.
Different letters above bars indicate mean group sizes that differ significantly; generalized linear model with negative binomial distribution with site as a random effect and species as a fixed effect (E. fuscus, n = 202 observed groups, intercept: 0.020 ± 0.11; M. lucifugus, n = 1,011 observed groups, coefficient 0.450 ± 0.08; M. septentrionalis, n = 151 observed groups, coefficient: −0.03 ± 0.12; P. subflavus, n = 263 observed groups, coefficient, −0.058 ± 0.11). The lower and upper hinges of the box plot show the first and third quartiles. The upper and lower whiskers extend to the largest and smallest value 1.5 times the interquartile range.
Extended Data Fig. 6 Distributions of shared-group sizes for four species of hibernating bats in early (November) and late (March) winter and during the collection of UVF-dust and fungal infection data.
a, Across all species, there was no significant change in social-group size over the winter using season (early versus late winter) as a fixed effect, and site and species as random effects (early winter, intercept: 0.38 ± 0.21; late winter, coefficient: 0.01 ± 0.03, P = 0.716, n = 1,956 bat groupings). We also investigated changes in social-group size for M. lucifugus, using an identical model but dropping species as a random effect. Again, we found no significant change in social group size over the winter (early winter, intercept: 0.84 ± 0.17, n = 457; late winter, coefficient: −0.008 ± 0.04, n = 528, P = 0.84). Neither model was significantly better than a null-intercept model. For E. fuscus, shared-group sizes increased slightly over the winter (early winter, intercept: 0.11 ± 0.11, n = 79; late winter, coefficient: 0.28 ± 0.14, n = 100, P = 0.040). b, We compared social-group size (n = 1,328) between the disease-arrival year and the dust-study year using a generalized linear mixed-effects model with a Poisson distribution and a log link. We found no significant difference in social-group sizes among years using study year as a categorical fixed effect (dust or disease) and site and species as random effects (disease study, intercept: 0.32 ± 0.16; UVF-dust study, coefficient: −0.04 ± 0.04, P = 0.33).
Extended Data Fig. 7 Environmental contamination and spatial overlap of UVF-dusted individuals.
a, Total surface area of the hibernacula walls and ceilings with UVF dust originating from different species (n = 51 measurements). Small black points show the summed total surface area with each colour of dust. Large points show the mean and 95% confidence intervals of the three species that were originally dusted in November (generalized linear mixed-effects model with site as a random effect and species as a fixed effect; M. lucifugus, intercept: 2.71 ± 0.15; M. septentrionalis, coefficient: 0.14 ± 0.17, t = 0.79; P. subflavus, coefficient: −0.21 ± 0.18, t = −1.16; species effect, P = 0.30). b, Percentage overlap of hibernacula areas used by individual bats compared to areas used by other individuals (n = 172 individual overlap estimates). The home range was calculated for each individual UVF-dust colour based on locations in the environment at which bats deposited their unique colour of UVF dust (see Methods). The points show an overlap estimate for each individual dusted bat compared to another dusted bat at that same site with larger points showing the mean and 95% confidence intervals for each species (generalized linear mixed-effects model with site as a random effect and species as a fixed effect; M. lucifugus, intercept: 0.88 ± 0.05; M. septentrionalis, coefficient: −0.03 ± 0.05, t = −0.50; P. subflavus, coefficient: −0.21 ± 0.07, t = −2.86; species effect, P = 0.02). Points are jittered in b to show overlapping data.
Extended Data Fig. 8 The probability of an individual being re-sighted with UVF dust by group size.
a, The probability of M. lucifugus (n = 1,697 individuals re-sighted for all dust colours) individuals having UVF dust that originated from another M. lucifugus increased with group size (generalized linear mixed-effects model with site and individual bat as random effects, and group size as a fixed effect; group size, coefficient: 0.017 ± 0.005; intercept, −0.596 ± 0.201, P = 0.0015). b, The probability of M. lucifugus (n = 4,164 individuals re-sighted for all dust colours) having UVF dust that originated from dusted M. septentrionalis decreased with M. lucifugus group size (group size coefficient, −0.013 ± 0.004; intercept, −1.973 ± 0.221, P = 0.0003). This suggests that M. septentrionalis were more likely to contact M. lucifugus roosting solitarily than in groups, whereas M. lucifugus roosting in groups were more likely to be contacted by other M. lucifugus than were M. lucifugus individuals that roosted solitarily.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion regarding conservation implications, Supplementary References and Supplementary Tables 1-9.
Rights and permissions
About this article
Cite this article
Hoyt, J.R., Langwig, K.E., White, J.P. et al. Cryptic connections illuminate pathogen transmission within community networks. Nature 563, 710–713 (2018). https://doi.org/10.1038/s41586-018-0720-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0720-z
Keywords
This article is cited by
-
Environmental transmission of Pseudogymnoascus destructans to hibernating little brown bats
Scientific Reports (2023)
-
Prevalence of Ophidiomyces ophidiicola and epizootiology of snake fungal disease in free-ranging Northern Pine Snakes (Pituophis melanoleucus melanoleucus) in New Jersey
Environmental Monitoring and Assessment (2023)
-
Human Exposure to Bats, Rodents and Monkeys in Bangladesh
EcoHealth (2023)
-
Ecology and impacts of white-nose syndrome on bats
Nature Reviews Microbiology (2021)
-
Group size and modularity interact to shape the spread of infection and information through animal societies
Behavioral Ecology and Sociobiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.