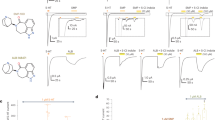
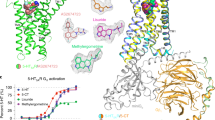
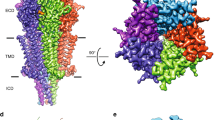
Abstract
The serotonin 5-HT3 receptor is a pentameric ligand-gated ion channel (pLGIC). It belongs to a large family of receptors that function as allosteric signal transducers across the plasma membrane1,2; upon binding of neurotransmitter molecules to extracellular sites, the receptors undergo complex conformational transitions that result in transient opening of a pore permeable to ions. 5-HT3 receptors are therapeutic targets for emesis and nausea, irritable bowel syndrome and depression3. In spite of several reported pLGIC structures4,5,6,7,8, no clear unifying view has emerged on the conformational transitions involved in channel gating. Here we report four cryo-electron microscopy structures of the full-length mouse 5-HT3 receptor in complex with the anti-emetic drug tropisetron, with serotonin, and with serotonin and a positive allosteric modulator, at resolutions ranging from 3.2 Å to 4.5 Å. The tropisetron-bound structure resembles those obtained with an inhibitory nanobody5 or without ligand9. The other structures include an ‘open’ state and two ligand-bound states. We present computational insights into the dynamics of the structures, their pore hydration and free-energy profiles, and characterize movements at the gate level and cation accessibility in the pore. Together, these data deepen our understanding of the gating mechanism of pLGICs and capture ligand binding in unprecedented detail.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates of the four conformations have been deposited in the Protein Data Bank with accession numbers 6HIN, 6HIO, 6HIQ and 6HIS for conformations F, I1, I2 and T, respectively. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession numbers EMD-0225, EMD-0226, EMD-0227 and EMD-0228 for conformations F, I1, I2 and T, respectively.
References
Thompson, A. J., Lester, H. A. & Lummis, S. C. R. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43, 449–499 (2010).
Nemecz, À., Prevost, M. S., Menny, A. & Corringer, P.-J. Emerging molecular mechanisms of signal transduction in pentameric ligand-gated ion channels. Neuron 90, 452–470 (2016).
Sparling, B. A. & DiMauro, E. F. Progress in the discovery of small molecule modulators of the Cys-loop superfamily receptors. Bioorg. Med. Chem. Lett. 27, 3207–3218 (2017).
Sauguet, L. et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl Acad. Sci. USA. 111, 966–971 (2014).
Hassaïne, G. et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281 (2014).
Miller, P. S. & Aricescu, A. R. Crystal structure of a human GABAA receptor. Nature 512, 270–275 (2014).
Du, J., Lü, W., Wu, S., Cheng, Y. & Gouaux, E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229 (2015).
Morales-Perez, C. L., Noviello, C. M. & Hibbs, R. E. X-ray structure of the human α4β2 nicotinic receptor. Nature 538, 411–415 (2016).
Basak, S. et al. Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat. Commun. 9, 514 (2018).
Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 346, 967–989 (2005).
Mukhtasimova, N., Lee, W., Wang, H. & Sine, S. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature 459, 451–454 (2009).
Lape, R., Colquhoun, D. & Sivilotti, L. G. On the nature of partial agonism in the nicotinic receptor superfamily. Nature 454, 722–727 (2008).
Menny, A. et al. Identification of a pre-active conformation of a pentameric channel receptor. eLife 6, e23955 (2017).
Corradi, J., Gumilar, F. & Bouzat, C. Single-channel kinetic analysis for activation and desensitization of homomeric 5-HT3A receptors. Biophys. J. 97, 1335–1345 (2009).
Sakmann, B., Patlak, J. & Neher, E. Single acetylcholine-activated channels show burst-kinetics in presence of desensitizing concentrations of agonist. Nature 286, 71–73 (1980).
Trick, J. L. et al. Functional annotation of ion channel structures by molecular simulation. Structure 6, 2207–2216 (2016).
Gasiorek, A. et al. Delineation of the functional properties and the mechanism of action of TMPPAA, an allosteric agonist and positive allosteric modulator of 5-HT3 receptors. Biochem. Pharmacol. 110–111, 92–108 (2016).
Jin, X. & Steinbach, J. H. A portable site: a binding element for 17β-estradiol can be placed on any subunit of a nicotinic α4β2 receptor. J. Neurosci. 31, 5045–5054 (2011).
Mukhtasimova, N. & Sine, S. M. Nicotinic receptor transduction zone: invariant arginine couples to multiple electron-rich residues. Biophys. J. 104, 355–367 (2013).
Cymes, G. D. & Grosman, C. The unanticipated complexity of the selectivity-filter glutamates of nicotinic receptors. Nat. Chem. Biol. 8, 975–981 (2012).
Zhu, F. & Hummer, G. Drying transition in the hydrophobic gate of the GLIC channel blocks ion conduction. Biophys. J. 103, 219–227 (2012).
Aryal, P., Sansom, M. S. P. & Tucker, S. J. Hydrophobic gating in ion channels. J. Mol. Biol. 427, 121–130 (2015).
Gielen, M., Thomas, P. & Smart, T. G. The desensitization gate of inhibitory Cys-loop receptors. Nat. Commun. 6, 6829 (2015).
McKinnon, N. K., Bali, M. & Akabas, M. H. Length and amino acid sequence of peptides substituted for the 5-HT3A receptor M3M4 loop may affect channel expression and desensitization. PLoS ONE 7, e35563 (2012).
Pless, S. A., Dibas, M. I., Lester, H. A. & Lynch, J. W. Conformational variability of the glycine receptor M2 domain in response to activation by different agonists. J. Biol. Chem. 282, 36057–36067 (2007).
Dahan, D. S. et al. A fluorophore attached to nicotinic acetylcholine receptor βM2 detects productive binding of agonist to the αδ site. Proc. Natl Acad. Sci. USA 101, 10195–10200 (2004).
daCosta, C. J. B. & Baenziger, J. E. Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure 21, 1271–1283 (2013).
Gonzalez-Gutierrez, G., Wang, Y., Cymes, G. D., Tajkhorshid, E. & Grosman, C. Chasing the open-state structure of pentameric ligand-gated ion channels. J. Gen. Physiol. 149, 1119–1138 (2017).
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011).
Hassaïne, G. et al. Large scale expression and purification of the mouse 5-HT3 receptor. BBA Biomembranes 1828, 2544–2552 (2013).
Hassaïne, G. et al. Expression, biochemistry, and stabilization with camel antibodies of membrane proteins: case study of the mouse 5-HT3 receptor. Methods Mol. Biol. 1635, 139–168 (2017).
Yakel, J. L., Lagrutta, A., Adelman, J. P. & North, R. A. Single amino acid substitution affects desensitization of the 5-hydroxytryptamine type 3 receptor expressed in Xenopus oocytes. Proc. Natl Acad. Sci. USA 90, 5030–5033 (1993).
Hu, X.-Q. & Lovinger, D. M. Role of aspartate 298 in mouse 5-HT3A receptor gating and modulation by extracellular Ca2+. J. Physiol. (Lond.) 568, 381–396 (2005).
Biyani, N. et al. Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Terwilliger, T. C., Sobolev, O. V., Afonine, P. V. & Adams, P. D. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D 74, 545–559 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Tuckerman, M., Berne, B. J. & Martyna, G. J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 97, 1990–2001 (1992).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Andersen, H. C. Rattle: A ‘velocity’ version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys. 52, 24–34 (1983).
Miyamoto, S. & Kollman, P. A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Comer, J., Phillips, J. C., Schulten, K. & Chipot, C. Multiple-replica strategies for free-energy calculations in NAMD: multiple-walker adaptive biasing force and walker selection rules. J. Chem. Theory Comput. 10, 5276–5285 (2014).
Comer, J. et al. The adaptive biasing force method: everything you always wanted to know but were afraid to ask. J. Phys. Chem. B 119, 1129–1151 (2015).
Moreau, C. J., Niescierowicz, K., Caro, L. N., Revilloud, J. & Vivaudou, M. Ion channel reporter for monitoring the activity of engineered GPCRs. Methods Enzymol. 556, 425–454 (2015).
Yang, J. Ion permeation through 5-hydroxytryptamine-gated channels in neuroblastoma N18 cells. J. Gen. Physiol. 96, 1177–1198 (1990).
Trattnig, S. M. et al. Discovery of a novel allosteric modulator of 5-HT3 receptors: inhibition and potentiation of Cys-loop receptor signaling through a conserved transmembrane intersubunit site. J. Biol. Chem. 287, 25241–25254 (2012).
Suryanarayanan, A. et al. The loop C region of the murine 5-HT3A receptor contributes to the differential actions of 5-hydroxytryptamine and m-chlorophenylbiguanide. Biochemistry 44, 9140–9149 (2005).
Yan, D., Schulte, M. K., Bloom, K. E. & White, M. M. Structural features of the ligand-binding domain of the serotonin 5HT3 receptor. J. Biol. Chem. 274, 5537–5541 (1999).
Ruepp, M.-D., Wei, H., Leuenberger, M., Lochner, M. & Thompson, A. J. The binding orientations of structurally-related ligands can differ; a cautionary note. Neuropharmacology 119, 48–61 (2017).
Kesters, D. et al. Structural basis of ligand recognition in 5-HT3 receptors. EMBO Rep. 14, 49–56 (2013).
Hibbs, R. E. et al. Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal α7 nicotinic acetylcholine receptor. EMBO J. 28, 3040–3051 (2009).
Spurny, R. et al. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl Acad. Sci. USA 109, E3028–E3034 (2012).
Delbart, F. et al. An allosteric binding site of the α7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein. J. Biol. Chem. 293, 2534–2545 (2018).
Steward, L. J. et al. Importance of phenylalanine 107 in Agonist recognition by the 5-hydroxytryptamine3A receptor. Mol. Pharmacol. 57, 1249–1255 (2000).
Purohit, P. & Auerbach, A. Acetylcholine receptor gating at extracellular transmembrane domain interface: the ‘pre-M1’ linker. J. Gen. Physiol. 130, 559–568 (2007).
Lee, W. Y. & Sine, S. M. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature 438, 243–247 (2005).
Hu, X.-Q., Zhang, L., Stewart, R. R. & Weight, F. F. Arginine 222 in the pre-transmembrane domain 1 of 5-HT3A receptors links agonist binding to channel gating. J. Biol. Chem. 278, 46583–46589 (2003).
Acknowledgements
We acknowledge access to the C-CINA and ESRF Krios microscopes, and thank M. Chami, L. Kovacik, H. Stahlberg, G. Effantin, E. Kandiah and M. Hons for support. We thank L. Estrozi, M. Bacia, G. Effantin and A. Desfosses for advice on cryo-EM; the Vivaudou and Moreau laboratories for providing high-quality oocytes and help with electrophysiology; members of the Nury laboratory, M. Gielen and E. Pebay-Peyroula for discussions. The work was funded by the Marie Curie CIG NeuroPenta and ERC Starting grant 637733 (to H.N.). It used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10 INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble PSB. The electron microscopy facility is supported by the Rhône-Alpes Region, the FRM, the FEDER and the GIS-IBISA.
Reviewer information
Nature thanks S. Sine, A. Sobolevsky and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
L.P., J.P. and G.H. performed sample preparation. L.P., H.N, E.N. and G.S. optimized and collected microscopy data. L.P. and H.N. built the models. F.D. and C.C. conducted molecular dynamics simulations. J.N. performed electrophysiology experiments. S.N.L. and P.-J.C. performed VCF experiments. A.A.J. performed FLIPR experiments. All authors extensively discussed the data, H.N. and J.N. wrote the manuscript with inputs from all authors.
Corresponding authors
Ethics declarations
Competing Interests
G.H. is employed by Theranyx. The remaining authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 5-HT3 receptor topology and sequence.
a, Schematic of the full-length 5-HT3 receptor. The colour code is the same for all panels. Scissors indicate the enzymatic treatments used during purification: purification tag removal using TEV protease, and partial removal of carbohydrates using PNGase F. b, Topology scheme and structure of one subunit of the 5-HT3 receptor. c, Sequence and numbering of the mouse 5-HT3A receptor used in the present study. Secondary structures and important loops are indicated. Neurotransmitter site binding loops are highlighted. Important residues discussed in the study are boxed (pore-facing in red, sandwich motif in brown and neurotransmitter site in blue). Cyan highlights residues important for TMPPAA potency. Glycosylation sites are depicted in green. The mouse receptor used for this study is a variant compared to the consensus sequence as it contains an alanine insertion in the M2–M3 loop, which is highlighted by the pink box. Notably, this insertion is present in the consensus human receptor sequence. This panel is adapted from ref. 5. d, A typical unmasked reconstruction, with different density levels overlaid depicting the protein itself (yellow), ligands (purple), linked glycans (green), the detergent-lipid belt (transparent purple) and the footprint of the disordered zone (transparent grey). e, Currents induced in Xenopus oocytes expressing the mouse 5-HT3A receptor by a 3-min application of 10 µM serotonin (green traces), co-application of 10 µM serotonin and 100 nM tropisetron (following a 5-min pre-application of tropisetron, red trace), co-application of 10 µM serotonin and 10 µM of TMPPAA (following a 5-min pre-application of TMPPAA, black trace). Current traces are representative of 3 independently repeated experiments.
Extended Data Fig. 2 Electron microscopy and 3D reconstructions for the tropisetron dataset (T), and for the serotonin + TMPPAA dataset (I2).
a, Tropisetron dataset (T). Selection of 2D class averages from the set of particles used for refinement of the tropisetron reconstruction. b, Three-dimensional reconstruction from the tropisetron dataset, filtered and coloured according to RELION local resolution. A global and a cut-through side view are shown. c, FSC curves for the density map before and after RELION post-processing, and between the model and the final map. d, Serotonin + TMPPAA dataset (I2). Selection of good 2D classes after one round of classification yields a set of 126,000 particles. One round of 3D classification with no symmetry imposed yields one class with pentameric symmetry, amounting to 62,000 particles. e, Unmasked unsharpened refined 3D reconstruction, filtered and coloured according to RELION local resolution. The colour range for resolution is similar to the equivalent representations for the other datasets in Extended Data Fig. 4c, allowing direct comparison. f, FSC curves for the density map before and after RELION post-processing, and between the model and the final map. g, Left, a non-sharpened map filtered at 3.9 Å resolution, obtained with a subset of 55,000 particles from the I2 dataset, selected after further 3D classification focused on the ICD. The hypothetical trajectory of the polypeptide chain after MX is bordered by a green line. No full connectivity can be visualized and the model was not built. The chain appears to descend abruptly after MX, and may interact with MA of the neighbouring subunit at the level of H411, where there is a clear density. Then it must link to the beginning of MA, but maps show no information how it may do so. The schematic representation highlights that this putative trajectory contacts MA on the neighbouring subunit close to residue H411. Right, the corresponding sharpened and masked reconstruction, showing that model building is not possible. The density of the disordered stretch closed to H411 is highlighted by the pink ellipses.
Extended Data Fig. 3 Serotonin and tropisetron bound in the orthosteric site; the vestibule allosteric site.
a, Organization of one of the five equivalent binding sites at the subunit interfaces in the ECD. Binding loops A–C are located on the principal subunit, and binding elements D–G are on the complementary subunit. The density around serotonin in the I2 reconstruction is shown as a transparent blue surface. b, c, Serotonin (ball-and-stick) in the orthosteric site in the I2 structure. Aromatic residues (dotted surfaces in c), hydrogen bonds with main chain atoms (dashed lines) and charged residues within salt-bridge distances (D202 on the principal side, R65 on the complementary side) are noted in two orthogonal views. Of note, mutants D202A64 and R65A65 exhibit impaired serotonin binding with increases of around 140-fold and 50-fold, respectively, in Ki (the equilibrium inhibitor constant) during competition assay for [3H]-granisetron binding. d, Superimposition of tropisetron-bound T (blue) and serotonin-bound I2 (green) structures, highlighting loop C motion and quaternary reorganization with arrows (principal subunit superposition in this panel and in g and h; note complementary subunit β-strand shift). Serotonin (light green) and tropisetron (magenta) are represented as sticks. e, f, Tropisetron bound to the orthosteric site in the T structure. The bicyclic tropane moiety is sandwiched between W156 of loop B and Y207 of loop C, whereas the indole lies close to R65, W63 in loop D, D42 and I44 in loop G, and R169 in loop F. Functional exploration of the binding mode of tropisetron consistently showed that, among others, single mutations to cysteine of the aromatic residues W156, Y207 or W63 abolished binding66. To fit tropisetron, the side chain of R65 is pushed upward and in turn displaces the side chains of Y67 and W168, compared to the empty orthosteric site of the X-ray structure. Densities for these side chains are not well-resolved. The density for tropisetron is shown in Extended Data Fig. 5f. g, Superposition of the I2 structure with the crystal structure of a mutant acetylcholine binding protein (AChBP, wheat cartoon) in complex with serotonin67 (yellow sticks) shows distinct binding modes. h, Superposition of the T structure with the crystal structure of AChBP (wheat cartoon) in complex with tropisetron (yellow sticks) shows a similar orientation of ligands but different interactions with the protein at loop C, D, E, F and G (not shown for clarity), consistent with the 3 orders of magnitude difference (0.7 versus 479 nM68) in Kd (dissociation constant). i, Overlay of the T and I2 structures (in blue and green, respectively) superimposed on a subunit ECD, showing the motion of F103 out of the vestibule site. The protein depicted as ribbon is viewed from inside the vestibule, and the intra-subunit cavity (in the I2 conformation) is represented as a grey surface. Acetate in GLIC, flurazepam in the bacterial homologue ELIC69 or a drug fragment in α7-AChBP70 bind to this cavity. A putative rationale for the strong effect of F103 mutants on serotonin EC5071, despite its absence of participation in the binding site, comes from the observation of its concerted motion with that of strands from the neighbouring subunit (around P128 and around P110; for clarity, the latter is not represented here). Therefore, the motion of F103 may participate in subunit–subunit quaternary reorganization, exemplifying the allosteric coupling between sites.
Extended Data Fig. 4 Electron microscopy, classifications and 3D reconstructions for the serotonin dataset.
a, Schematic of the data processing, with classical rounds of 2D and 3D classification yielding a consensus reconstruction seen parallel to the membrane plane (grey) where helices look like tubes of elliptic sections, indicating heterogeneity. Extensive classification trials, without alignment, with masks of various size covering the lower part of the TMD (shown in green and red) enable two conformations to be distinguished (yellow corresponding to the I1 structure, and purple corresponding to the F structure), representing the extreme positions of the helices in the consensus reconstruction. An overlay of the two reconstructions depicts the good superimposition of the ECD and the clear difference in the lower TMD. Grey rectangles indicate the positions of the slabs represented in b. All reconstructions shown are unmasked outputs of RELION 3D refinements. b, Slabs, viewed perpendicular to the membrane plane, from the intracellular side showing the overlay of the I1 and the F reconstructions. c, Reconstructions filtered and coloured according to RELION local resolution. A global and a cut-through side view are shown. d, Selection of 2D class averages from the set of particles used for refinement of the I1 and F reconstructions. e, FSC curves showing the unmasked and masked FSCs (before and after post-processing in RELION), and the model-to-map FSC. f, Selection of ‘slice’ views of the TMD of the final unmasked unsharpened reconstructions.
Extended Data Fig. 5 Quality of the density maps in representative regions of the four 5-HT3 receptor reconstructions.
Densities in mesh representations overlaid with structures for the T (blue), I1 (yellow), F (purple) and I2 (green) reconstructions and structures, left to right, respectively, in each panel. Views a–e are approximately the same as those in extended data figure 4 of a previous publication of α1 GlyR7. a, Representative densities of the β-sheets in the ECD. b, Densities of the Cys loop and the M2–M3 loop. c, Densities of helices M1 and M2. d, Densities of M3 and M4. e, Densities of M2 at the level of L9′ (L260). f, Densities around the ligands tropisetron (in T) and serotonin (in I2). The 3.2 Å resolution of I2 permits unambiguous orientation of serotonin, and side chains of residues around the ligands are seen in densities. For tropisetron, given the limited 4.5 Å global resolution, the orientation of the ligand and the positions of surrounding side chain are less certain. The views are the same as in Extended Data Fig. 3b, e.
Extended Data Fig. 6 Structural superpositions with the nAChR α4β2, the GABAA receptor β3 subunit and the apo 5-HT3 receptor structures.
a, Putty representations of pairwise deviations for the 5-HT3 receptor conformations. The selection used for superimposition, and the two conformations used, are noted for each image. The colour code and tube thickness code are the same for all images. In the T versus X-ray image, the red zone corresponds to a loop that was not modelled in the X-ray structure. b, Superimposition of a 5-HT3 receptor subunit in the inhibited T state (blue) and of a 5-HT3 receptor subunit in the apo state (grey, PDB: 6BE1). Structures are globally similar with differences in the lipid-exposed helices M3, MX and M4, hypothetically a consequence of the different additives used—a lipid mixture in this study versus fluorinated fos-choline 8 for the apo structure. Superposition of the T structure (blue), the apo structure (grey) and the X-ray structure (orange). c, Superposition of a 5-HT3 receptor subunit in the I1, I2 and F conformations with a nAChR α4 subunit (PDB: 5KXI, chain A) or a GABAA receptor β3 subunit (PDB: 4COF). Cα r.m.s.d. is noted. Comparison of superpositions with r.m.s.d. below 2 Å shows that in the case of F and nAChR α4 (middle, purple and grey), the domain-to-domain orientation is very similar and the extracellular halves of helices M1, M2, M3 superimpose very well, whereas clear deviations are present in their intracellular halves and at the level of M4 (r.m.s.d. without M4 drops to 1.6 Å). In the case of I1 and GABAA β3 (left, yellow and black), or I2 and GABAA β3 (right, green and black), differences are more distributed; deviations of the ECD indicate different domain-to-domain orientations, and in the TMD M1 and M3 superimpose quite well but M2 and M4 clearly differ.
Extended Data Fig. 7 TMPPAA binds to an allosteric site in the TMD.
a, Cavities in surface mode for the T (blue), I1 (yellow), F (purple) and I2 (green) states. Note the re-arrangement of the cavities between states. The protein is depicted in cartoon, with sticks in the right panels for residues in which mutations impair the effect of TMPPAA. Noisy densities are found in the I2 reconstruction, in or close to the cavities, but they do not permit ligand modelling and could correspond to parts of a lipid or detergent molecule. b, Side view of the transmembrane domain of I2 with mutated positions depicted as green spheres (for residues for which mutations had negligible effect on TMPPAA potency), orange spheres (for residues for which mutations reduced TMPPAA potency substantially) or red spheres (for mutations that completely or almost completely eliminated TMPPAA activity at concentrations up to 100 µM). Yellow circles indicate residues for which different mutations produced different effects. c, Functional properties exhibited by serotonin and TMPPAA as agonists at 5-HT3A receptors expressed in tsA201 cells in an FMP assay. The human 5-HT3A receptor was used for these experiments, since TMPPAA evokes a more robust agonist response through this receptor than through mouse 5-HT3A in this assay17. The colour code is similar to that in b. n.d., not determinable, w.a., weak activity. A complete table of data for all tested mutant receptors is shown in Supplementary Table 1. d, Concentration-response curves for serotonin (closed symbols) and TMPPAA (open symbols) tested as agonists at wild-type, W472A and L243W human 5-HT3A receptors expressed in tsA201 cells, using the FMP assay. Data are from a single representative experiment determined in the same 96-well plate and are given as mean based on duplicate determinations. The experiment was repeated independently with similar results 3 times for the W472A and L243W receptors and 66 times for the wild-type receptor (n for wild-type and all mutant receptors are given in c). e, Loss of TMPPAA potency at the mouse W456A mutant receptor. Currents evoked by 10 µM serotonin (blue curve) or 10 µM serotonin plus 100 µM TMPPAA (red curve) are equivalent. The grey trace, obtained in an oocyte expressing wild-type receptors (response to 10 µM serotonin, normalized for the peak amplitude), shows that desensitization in wild-type and W456A mutant receptors is similar. Current traces are representative of 3 independently repeated experiments.
Extended Data Fig. 8 Structural motif and reorganization at the ECD–TMD interface.
a, b, Key interacting charged residues R218 (pre-M1, key in gating72,73,74), E53 (loop β1–β2), E186 (loop β8–β9, conserved only in cationic eukaryotic receptors) and D145 (Cys loop)—represented as purple sticks—are sandwiched between conserved essential aromatic residues (FPF motif of the Cys loop, W187 of β8–β9 and Y223 of M1), represented as yellow sticks and dots. Lower in the membrane, the strictly conserved P230—represented as spheres—enables M1 to kink. This structural organization is common to all pLGICs of known structure. Orientations of the views are indicated on the topology scheme. c, d, Side and top views depicting the concerted relative positions of the Cys-loop FPF ‘wedge’, represented as sticks, of transmembrane helices and of the β8–β9 loop. Compared to the inhibited structure, the wedge moves towards and pushes on M1 and also moves towards M4 in I1, I2 and F. F presents the biggest re-organization in this zone: M1, the M2–M3 loop and M3 follow the motion of the FPF motif, and M2 moves away from the pore axis (that is, to the back in the view in c and to the top in d). e, Conservation of the ‘sandwich’ structural motif in representative pLGIC structures. The aromatic top and bottom layers are represented as spheres (yellow, Cys loop; orange, β8–β9 loop and pre-M1 residue) whereas the central layer of charges is depicted as sticks. Note that in anionic receptors and GLIC, the charge of β8–β9 is absent (noted with a star). f, Sequence alignment of the motif residues in representative pLGIC structures.
Extended Data Fig. 9 Geometry of the pore during the molecular dynamics simulations.
a, HOLE transmembrane pore profiles for the trajectories starting from I1, F and I2. In each graph, the profile from the cryo-EM structure is represented in black, the average profile during the restrained part of the simulation (30–60 ns, mean ± s.d. of n = 30 snapshots taken every ns) in the lighter colour and the average profile during the unrestrained part (60–1,000 ns, mean ± s.d. of n = 940 snapshots taken every ns) in the darker colour. The y-axis origin corresponds to L260/9′ whereas S253/2′s is located at about −12 Å. b, Complete potentials of mean force of K+ ions as a function of position along the pore axis. The inset represents the 2D free-energy landscape as a function of the position along the pore axis and the radial direction, orthogonal to it. The represented free-energy profile is the mean of 135,000,000 values distributed along 2.16 µs of the I1 trajectory (190,000,000 values distributed along 3.04 µs of the F trajectory); the standard deviation corresponds in both cases to 50,000,000 values from four independent walkers, distributed along the last 0.8 µs of the trajectory. c, Representative snapshots of the pore wetting during the initial restrained part (30–60 ns) of the simulations. A dewetted pore is observed for the T and I1 trajectories, a fully wetted one for the F trajectory, whereas wetting and dewetting occur in the I2 trajectory. Cα atoms of pore-exposed residues are represented as spheres. d, Snapshot of the I2 trajectory showing the wetting of grooves at the back of M2 (shown as molecular surface on the left, as cartoon on the right), concomitant with wetting of the pore.
Extended Data Fig. 10 Local motions in the pore probed by VCF.
Representative serotonin-evoked simultaneous current and fluorescence recordings from oocytes expressing S19′C mutant receptors labelled with MTS-TAMRA. Similar traces were obtained in 3 independent experiments.
Supplementary information
Supplementary Table
This file contains SI Table 1: Functional properties displayed by 5-HT and TMPPAA tested as agonists at tsA201 cells transiently transfected with wild-type and mutant human 5-HT3A receptors in the FLIPR Membrane Potential Blue assay. SI Table 2: Oligonucleotides used for the study.
Video 1: T→I1→F morph, side view
Superimposition on ECDs. Cartoon representation of the pentamer viewed parallel to the membrane plane. Colours follow the scheme used throughout the paper T blue, I1 yellow, F purple.
Video 2: T→I1→F morph, top view of the TMD
Superimposition on ECDs. Cartoon representation of the pentamer TMD viewed perpendicular to the membrane plane. T blue, I1 yellow, F purple.
Video 3: T→I1→F morph
Superimposition on the TMD of one subunit. Cartoon representation of a subunit viewed parallel to the membrane plane. T blue, I1 yellow, F purple.
Video 4: Ions motions through the pore in the F trajectory
Two M2 helices are represented as purple cartoon with C of pore-exposed residues as white spheres. K+ ions in the pore region are depicted as colour spheres.
Rights and permissions
About this article
Cite this article
Polovinkin, L., Hassaine, G., Perot, J. et al. Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279 (2018). https://doi.org/10.1038/s41586-018-0672-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0672-3
Keywords
This article is cited by
-
Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment
Nature Communications (2024)
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
A release of local subunit conformational heterogeneity underlies gating in a muscle nicotinic acetylcholine receptor
Nature Communications (2024)
-
Structural basis for partial agonism in 5-HT3A receptors
Nature Structural & Molecular Biology (2024)
-
α7 Nicotinic acetylcholine receptor: a key receptor in the cholinergic anti-inflammatory pathway exerting an antidepressant effect
Journal of Neuroinflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.