Abstract
Skeletal stem cells regulate bone growth and homeostasis by generating diverse cell types, including chondrocytes, osteoblasts and marrow stromal cells. The emerging concept postulates that there exists a distinct type of skeletal stem cell that is closely associated with the growth plate1,2,3,4, which is a type of cartilaginous tissue that has critical roles in bone elongation5. The resting zone maintains the growth plate by expressing parathyroid hormone-related protein (PTHrP), which interacts with Indian hedgehog (Ihh) that is released from the hypertrophic zone6,7,8,9,10, and provides a source of other chondrocytes11. However, the identity of skeletal stem cells and how they are maintained in the growth plate are unknown. Here we show, in a mouse model, that skeletal stem cells are formed among PTHrP-positive chondrocytes within the resting zone of the postnatal growth plate. PTHrP-positive chondrocytes expressed a panel of markers for skeletal stem and progenitor cells, and uniquely possessed the properties of skeletal stem cells in cultured conditions. Cell-lineage analysis revealed that PTHrP-positive chondrocytes in the resting zone continued to form columnar chondrocytes in the long term; these chondrocytes underwent hypertrophy, and became osteoblasts and marrow stromal cells beneath the growth plate. Transit-amplifying chondrocytes in the proliferating zone—which was concertedly maintained by a forward signal from undifferentiated cells (PTHrP) and a reverse signal from hypertrophic cells (Ihh)—provided instructive cues to maintain the cell fates of PTHrP-positive chondrocytes in the resting zone. Our findings unravel a type of somatic stem cell that is initially unipotent and acquires multipotency at the post-mitotic stage, underscoring the malleable nature of the skeletal cell lineage. This system provides a model in which functionally dedicated stem cells and their niches are specified postnatally, and maintained throughout tissue growth by a tight feedback regulation system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source Data are provided in the online version of the paper. The datasets generated during and/or analysed during the current study are available in Dryad Digital Repository (https://doi.org/10.5061/dryad.3qq5bm7).
References
Ono, N. & Kronenberg, H. M. Bone repair and stem cells. Curr. Opin. Genet. Dev. 40, 103–107 (2016).
Ono, N., Ono, W., Nagasawa, T. & Kronenberg, H. M. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 16, 1157–1167 (2014).
Chan, C. K. et al. Identification and specification of the mouse skeletal stem cell. Cell 160, 285–298 (2015).
Worthley, D. L. et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell 160, 269–284 (2015).
Kronenberg, H. M. Developmental regulation of the growth plate. Nature 423, 332–336 (2003).
St-Jacques, B., Hammerschmidt, M. & McMahon, A. P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 (1999).
Kobayashi, T. et al. PTHrP and Indian hedgehog control differentiation of growth plate chondrocytes at multiple steps. Development 129, 2977–2986 (2002).
Kobayashi, T. et al. Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J. Clin. Invest. 115, 1734–1742 (2005).
Chen, X. et al. Initial characterization of PTH-related protein gene-driven lacZ expression in the mouse. J. Bone Miner. Res. 21, 113–123 (2006).
Mak, K. K., Kronenberg, H. M., Chuang, P. T., Mackem, S. & Yang, Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development 135, 1947–1956 (2008).
Abad, V. et al. The role of the resting zone in growth plate chondrogenesis. Endocrinology 143, 1851–1857 (2002).
Ara, T. et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J. Immunol. 170, 4649–4655 (2003).
Yang, L., Tsang, K. Y., Tang, H. C., Chan, D. & Cheah, K. S. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl Acad. Sci. USA 111, 12097–12102 (2014).
Bianco, P. et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 19, 35–42 (2013).
Bianco, P. ‘Mesenchymal’ stem cells. Annu. Rev. Cell Dev. Biol. 30, 677–704 (2014).
Pardo-Saganta, A. et al. Parent stem cells can serve as niches for their daughter cells. Nature 523, 597–601 (2015).
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014).
Busch, K. et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015).
Acknowledgements
We thank D. Holcomb and M. Curtis of Carl Zeiss Microscopy for assistance in imaging, G. Gavrilina and W. Fillipak of the University of Michigan Transgenic Animal Model Core for assistance with transgenesis. This research was supported by NIH R01DE026666 and R00DE022564 (to N.O.), R03DE027421 (to W.O.), P01DK011794 (to H.M.K.), 2017 Fred F. Schudy Memorial Research Award from the American Association of Orthodontists Foundation (to N.O.) and University of Michigan MCubed 2.0 Grant (to N.O. and W.O.).
Reviewer information
Nature thanks O. Klein, M. T. Longaker and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
K.M. and N.O. conceived the project and designed the experiments; K.M. and N.O. performed the mouse genetic experiments with assistance from W.O., N.S. and A.T., who performed genotyping; K.M. performed histological experiments and imaging analysis; K.M. performed cell culture experiments; K.M. and N.O. performed flow cytometry experiments and analysis; Y.M. performed the surgery and cell transplantation; K.M. and N.O. analysed the data; N.O. supervised the project; T.L.S. generated the mice; T.N. provided the mice; K.M. and N.O. wrote the manuscript; T.N., W.O. and H.M.K. critiqued the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
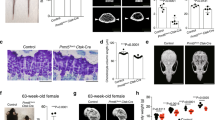
Extended Data Fig. 1 Generation and characterization of Pthrp-mCherry knock-in allele.
a, CRISPR–Cas9 generation of Pthrp-mCherry knock-in allele. Structure of the genomic Pthrp locus, targeting vector and knock-in allele after homologous recombination. White boxes, untranslated region; black boxes, coding region; ex, exon. Blue bars, homology arms; red bars, guide RNAs (gRNAs) as part of CRISPR–Cas9 reagents; red boxes, Kozak-mCherry-bGHpA cassette replacing the native start codon. Half arrows, primers; wild-type forward (289), wild-type reverse (290) and mutant reverse (291). Bottom, PCR genotyping using 289, 290 and 291 primer mix; wild-type (WT) allele, 185 bp; knock-in (KI) allele, 385 bp. At least n = 100 independent experiments with similar results. b, PthrpmCherry/+ fetal distal femurs with EdU administration shortly before analysis (3 h). Bottom panels show magnified views of perichondrium. Dotted lines, borders of bone anlage. Grey, DAPI and DIC. Scale bars, 200 μm (top panels), 100 μm (bottom panels). n = 2 (E13.5, E15.5) mice, n = 1 (α-Sox9) mouse. c, PthrpmCherry/+ distal-femur growth plates with EdU administration shortly before analysis (3 h). Bottom panels show magnified views of central growth plates. Arrowheads, mCherry+ cells. Grey, DAPI and DIC. Scale bars, 200 μm (top panels), 50 μm (bottom panels).
Extended Data Fig. 2 Skeletal stem and progenitor cell-marker expression in PTHrP–mCherry+ resting chondrocytes.
a, Flow cytometry analysis of PthrpmCherry/+ growth-plate cells (top panels) and bone-marrow cells (bottom panels). n = 8 mice for PthrpmCherry/+ and n = 3 mice for Pthrp+/+, data are presented as mean ± s.d. b, Flow cytometry analysis of Col1a1(2.3kb)-GFP;PthrpmCherry/+ growth-plate cells. n = 5 mice per group, data are presented as mean ± s.d. c, Skeletal stem and progenitor cell-surface-marker analysis of PthrpmCherry/+ growth-plate cells. Unstained, Pthrp+/+ cells mice only stained for CD45, Ter119 and CD31; mCherry−, mCherry− fraction of PthrpmCherry/+ cells; mCherry+, mCherry+ fraction of PthrpmCherry/+ cells. Magenta box, CD45−Ter119−CD31−CD51+CD90−mCherry+ fraction. n = 3 mice for PthrpmCherry/+, data are presented as mean ± s.d., n = 1 mouse for Pthrp+/+. d, Composition of CD45−Ter119−CD31−mCherry+ growth-plate cells. n = 3 mice per group, data are presented as mean ± s.d. e, Percentage of mCherry+ cells among mSSCs (left, CD105−CD200+), pre-BCSPs (centre, CD105−CD200−) and BCSPs (right, CD105+), gated under CD45−Ter119−CD31−CD51+CD90− fraction. n = 3 mice per group, data are presented as mean ± s.d.
Extended Data Fig. 3 Generation and characterization of Pthrp-creER bacterial artificial chromosome transgenic line.
a, Generation of Pthrp-creER bacterial artificial chromosome (BAC) transgenic mice. Structure of the Pthrp-creER-WPRE-rGHpA BAC construct. Kozak-Pthrp-creER-WPRE-rGHpA-frt-NeoR-frt cassette containing 62-bp homology arms was recombineered into a BAC clone RP23-27F7 containing 131-kb upstream and 82-kb downstream genomic sequences of the Pthrp gene. NeoR and backbone lox sites were removed before pronuclear injection. Half arrows, forward (62) and reverse (63) primers. Right, PCR genotyping using 62 and 63 primer mix; transgenic (Tg), 373 bp. White boxes, exons; black boxes, introns. At least n = 100 independent experiments with similar results. b, Short-chase analysis of Pthrp-creER;R26RZsGreen;PthrpmCherry/+ distal-femur growth plates (pulsed on P6). Scale bars, 50 μm. n = 3 mice. c–e, Short-chase flow cytometry analysis of Pthrp-creER;R26RZsGreen;PthrpmCherry/+ growth-plate cells, with tamoxifen injection at 72 h (c, e) or 22 h (d) in advance. Red lines, ZsGreen+ cells; blue lines, control cells without PTHrP–mCherry. n = 5 mice (72 h) or n = 3 mice (22 h) per group. e, Percentage of CD105+ cells within mCherry+ (red) and ZsGreen+ (green) cells. n = 5 mice per group, data are presented as mean ± s.d., *P = 0.012, Mann–Whitney’s U-test, two-tailed. f, Pthrp-creER;R26RtdTomato distal-femur growth plates (pulsed on P6) at P9. EdU (50 μg) was serially injected 9 times at 8-h intervals between P6 and P9. Grey, DIC. Scale bars: 50 μm. n = 3 mice. g, Scanning of Pthrp-creER;R26RtdTomato whole femur (pulsed on P6) at P12. Arrow, tdTomato+ cells localized within the resting zone of distal femur. Grey, DAPI and DIC. Scale bars, 1 mm. n = 3 mice. h, High sensitivity in situ hybridization (RNAscope) analysis of Pthrp-creER;R26RtdTomato distal-femur growth plates (pulsed on P6) at P12. Top and bottom panels represent the identical section, before (bottom panels) and after (top panels) hybridization. Left panels, Col2a1 (positive control); centre panels, Grem1; right panels, negative control. Grey, DAPI and DIC. Scale bars, 200 μm. n = 3 independent experiments.
Extended Data Fig. 4 PTHrP+ resting chondrocytes are functionally dedicated to columnar chondrocyte formation.
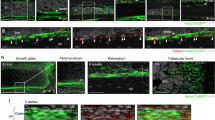
a, In vivo clonal analysis of Col2a1-creER+ growth-plate chondrocytes. Col2a1-creER;R26RConfetti distal-femur growth plates (pulsed on P6, P7 and P8). Scale bars, 50 μm. n = 2 mice. b–e, Col1a1(2.3kb)-GFP;Pthrp-creER;R26RtdTomato distal-femur growth plates, shown at P9 after being pulsed at various preceding time points. Yellow double-headed arrows, tdTomato+ domain within the resting zone. Grey, DAPI and DIC. Scale bars, 200 μm. n = 3 mice per group. f, g, Cxcl12-GFP;Pthrp-creER;R26RtdTomato distal-femur growth plates, shown at P36 after being pulsed on P0 (f) and P6 (g). Yellow double-headed arrows in f, g indicate the same width as those shown in d, e. Grey, DAPI and DIC. Scale bars, 200 μm. n = 3 mice per group.
Extended Data Fig. 5 Dlx5-creER+ proliferating chondrocytes are not the source of columnar chondrocytes.
a–d, Cell-fate analysis of Dlx5-creER+ proliferating chondrocytes. Dlx5-creER;R26RtdTomato distal-femur growth plates (pulsed on P7). b, EdU (200 μg) was serially injected 6 times at 8-h intervals, between P7 and P9. Arrows, EdU+tdTomato+ cells; arrowheads, short columns (<10 cells). Grey, DAPI and DIC. Scale bars, 200 μm (left panels), 50 μm (right panel). n = 3 mice at each time point.
Extended Data Fig. 6 Pthrp-creER+ resting chondrocytes are precursors for bone marrow reticular stromal cells.
Cxcl12-GFP;Pthrp-creER;R26RtdTomato distal femurs (pulsed on P6). a–d, Bottom panels show magnified views of the dotted areas beneath growth plates. Arrows, Cxcl12–GFP+tdTomato+ reticular stromal cells. e, Magnified view of the junction between hypertrophic layer and primary spongoisa. Arrow, Cxcl12–GFP+tdTomato+ reticular stromal cells immediately below the hypertrophic zone. 1°S: primary spongiosa. f, Magnified view of the metaphyseal bone marrow. Mice were fed with high-fat diet containing rosiglitazone between P56 and P97. Grey, DAPI and DIC. Scale bars, 500 μm (a–d, f), 100 μm (e), 50 μm (bottom panels of a–c), 20 μm (bottom panel of d, right panel of e). n = 3 mice for each group, except n = 2 mice for P365.
Extended Data Fig. 7 Pthrp-creER+ resting chondrocytes uniquely possess colony-forming capabilities ex vivo.
a, Diagram of colony-forming assay. Growth-plate cells were isolated from Pthrp-creER;R26RtdTomato (pulsed on P6) or Dlx5-creER;R26RtdTomato (pulsed on P7) mice at P9, and cultured at a clonal density (~1,000 cells per cm2) for 10–14 days to initiate colony formation. BM, bone marrow. b, Colony-forming assay. Left top, Pthrp-creER;R26RtdTomato; right, Dlx5-creER;R26RtdTomato. Insets 1,2 and 3 show magnified views of the corresponding areas (labelled with 1, 2, 3). Bottom left, Sox9 staining of primary Pthrp-creER tdTomato+ colonies. Red, tdTomato. Scale bars, 5 mm (top panels), 1 mm (top panel insets), 200 μm (bottom panel). n = 88 mice for Pthrp-creER;R26RtdTomato, n = 5 for Dlx5-creER;R26RtdTomato. c, Quantification of tdTomato+ colonies (>50 cells) established from Pthrp-creER;R26RtdTomato (n = 88) and Dlx5-creER;R26RtdTomato (n = 5) mice. Data are presented as mean ± s.d. d, Diagram of colony-forming assay and subsequent analyses on self-renewal, trilineage differentiation and transplantation of individual colony-forming cells. e, Isolation of single PTHrPCE–tdTomato+ colonies and subsequent subculture of isolated clones. A, exhausting clone; B, self-renewing clone establishing secondary colonies. Right, clone B did not proliferate at passage 2 upon bulk culture. Red, tdTomato. Scale bars, 5 mm. n = 518 independent experiments. f, Subcutaneous transplantation of PTHrPCE–tdTomato+ clones into immunodeficient mice. n = 8 mice.
Extended Data Fig. 8 Pthrp-creER+ resting chondrocytes form columnar chondrocytes in a Hedgehog-responsive, niche-dependent manner.
a–i, Pharmacological manipulation of Hedgehog signalling. Pthrp-creER;R26RtdTomato distal-femur growth plates (pulsed on P6). Left panels, vehicle control; centre panels, SAG (Hh agonist)-treated samples; right panels, LDE225 (Hh antagonist)-treated samples. Grey, DAPI and DIC. Scale bars, 200 μm. j, k, Quantification of tdTomato+ columns in Pthrp-creER;R26RtdTomato distal-femur growth plates (pulsed on P6). P17, n = 5 (control), n = 5 (SAG), n = 4 (LDE225) mice per group. P28, n = 4 (control), n = 3 (LDE225) mice per group. Data are presented as mean ± s.d. P28, n = 2 (SAG). ***P < 0.001; P17 control versus SAG, mean difference = 67.8, 95% confidence interval (37.5, 98.1); P17 control versus LDE225, mean difference = 66.0, 95% confidence interval (33.9, 98.0); P17 SAG versus LDE225, mean difference = −1.85, 95% confidence interval (−33.9, 30.2); P28 control versus LDE225, mean difference = 134.5, 95% confidence interval (108.7, 160.3). One-way ANOVA followed by Tukey’s multiple comparison test. l, m, Micro-perforation injury of growth plates. Col1a1(2.3kb)-GFP;Pthrp-creER;R26RtdTomato distal femurs (pulsed on P6) at P28. Micro-perforation surgery was performed at P21. l, Left femur growth plate (control). m, Right femur growth plate (micro-perforated). Dotted line, micro-perforated area. Grey, DAPI and DIC. Scale bars, 100 μm. n = 3 mice.
Extended Data Fig. 9 Resting zone of the growth plate contains a unique class of skeletal stem cells.
a, Formation of PTHrP+ skeletal stem cells within the growth plate. A small subset of PTHrP+ chondrocytes in the resting zone acquire properties as long-term skeletal stem cells in conjunction with the formation of the highly vascularized secondary ossification centre. b, PTHrP+ skeletal stem cells are heterogeneously composed of long-term, short-term and transient populations, and undergo asymmetric divisions and maintain themselves within the resting zone. These cells may be supplemented by PTHrP− cells. PTHrP+ cells perform two different functions: (1) these cells differentiate into proliferating chondrocytes, hypertrophic chondrocytes and eventually become osteoblasts and bone marrow stromal cells at the post-mitotic stage. (2) These cells send a forward signal (PTHrP) to control chondrocyte proliferation and differentiation. Indian hedgehog (Ihh) secreted by hypertrophic chondrocytes maintains the proliferation of chondrocytes and formation of columnar chondrocytes.
Extended Data Fig. 10 Absence of tamoxifen-independent recombination in Pthrp-creER line.
a, No tamoxifen controls of Pthrp-creER;R26RtdTomato mice at 6 months (left) and 1 year (right) of age. Red, tdTomato; blue, DAPI; grey, DIC. Scale bars, 500 μm. n = 3 mice per group. b, No tamoxifen controls of primary colonies (passage 0) isolated from Pthrp-creER;R26RtdTomato mice at P12 without tamoxifen injection. Left, methylene blue (MB) staining; right, red tdTomato (TOM). Scale bar, 5 mm. n = 3 mice. c, Dose–response curve of recombination based on Pthrp-creER. Quantification of tdTomato+ cells in resting zone at P9 in Pthrp-creER;R26RtdTomato mice upon a single dose of tamoxifen at P6. x axis, dose of tamoxifen (μg); y axis, the number of tdTomato+ cells per 1-mm thickness. n = 3 (0, 31.3 and 62.5 μg), n = 4 (15.6, 125, 250 and 500 μg) mice per group, data are presented as mean ± s.d. d, Tamoxifen-induced recombination in growth plates pulsed on P9. Pthrp-creER;R26RtdTomato distal-femur growth plates at P12 (left) and Col1a1(2.3kb)-GFP;Pthrp-creER;R26RtdTomato mice at P21 (right). Tamoxifen (500 μg) was injected at P9. Green, Col1a1(2.3kb)–GFP; red, tdTomato; grey, DAPI and DIC. Scale bars, 200 μm. n = 3 mice.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and References.
Rights and permissions
About this article
Cite this article
Mizuhashi, K., Ono, W., Matsushita, Y. et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature 563, 254–258 (2018). https://doi.org/10.1038/s41586-018-0662-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0662-5
Keywords
This article is cited by
-
Compensatory growth and recovery of cartilage cytoarchitecture after transient cell death in fetal mouse limbs
Nature Communications (2024)
-
Gli1 labels progenitors during chondrogenesis in postnatal mice
EMBO Reports (2024)
-
Piezo1 expression in chondrocytes controls endochondral ossification and osteoarthritis development
Bone Research (2024)
-
The emerging studies on mesenchymal progenitors in the long bone
Cell & Bioscience (2023)
-
Stem cells provide clues to why vertebrae attract tumour cells
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.