Abstract
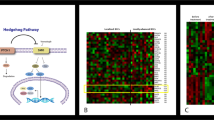
Basal cell carcinoma (BCC) is the most frequent cancer in humans and results from constitutive activation of the Hedgehog pathway1. Several Smoothened inhibitors are used to treat Hedgehog-mediated malignancies, including BCC and medulloblastoma2. Vismodegib, a Smoothened inhibitor, leads to BCC shrinkage in the majority of patients with BCC3, but the mechanism by which it mediates BCC regression is unknown. Here we used two genetically engineered mouse models of BCC4 to investigate the mechanisms by which inhibition of Smoothened mediates tumour regression. We found that vismodegib mediates BCC regression by inhibiting a hair follicle-like fate and promoting the differentiation of tumour cells. However, a small population of tumour cells persists and is responsible for tumour relapse following treatment discontinuation, mimicking the situation found in humans5. In both mouse and human BCC, this persisting, slow-cycling tumour population expresses LGR5 and is characterized by active Wnt signalling. Combining Lgr5 lineage ablation or inhibition of Wnt signalling with vismodegib treatment leads to eradication of BCC. Our results show that vismodegib induces tumour regression by promoting tumour differentiation, and demonstrates that the synergy between Wnt and Smoothened inhibitors is a clinically relevant strategy for overcoming tumour relapse in BCC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data associated with this study have been deposited in the NCBI Gene Expression Omnibus under accession number GSE117458 (microarray).
References
Epstein, E. H. Basal cell carcinomas: attack of the hedgehog. Nat. Rev. Cancer 8, 743–754 (2008).
Basset-Seguin, N., Sharpe, H. J. & de Sauvage, F. J. Efficacy of Hedgehog pathway inhibitors in basal cell carcinoma. Mol. Cancer Ther. 14, 633–641 (2015).
Sekulic, A. et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 366, 2171–2179 (2012).
Kasper, M., Jaks, V., Hohl, D. & Toftgård, R. Basal cell carcinoma - molecular biology and potential new therapies. J. Clin. Invest. 122, 455–463 (2012).
Tang, J. Y. et al. Inhibition of the hedgehog pathway in patients with basal-cell nevus syndrome: final results from the multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 17, 1720–1731 (2016).
Atwood, S. X. et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 27, 342–353 (2015).
Sharpe, H. J. et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 27, 327–341 (2015).
Youssef, K. K. et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat. Cell Biol. 12, 299–305 (2010).
Youssef, K. K. et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat. Cell Biol. 14, 1282–1294 (2012).
Brown, J. A. et al. TGF-β-induced quiescence mediates chemoresistance of tumor-propagating cells in squamous cell carcinoma. Cell Stem Cell 21, 650–664.e8 (2017).
Barker, N., Tan, S. & Clevers, H. Lgr proteins in epithelial stem cell biology. Development 140, 2484–2494 (2013).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Page, M. E., Lombard, P., Ng, F., Göttgens, B. & Jensen, K. B. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13, 471–482 (2013).
Yang, S. H. et al. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/β3-catenin signaling. Nat. Genet. 40, 1130–1135 (2008).
Rhee, H., Polak, L. & Fuchs, E. Lhx2 maintains stem cell character in hair follicles. Science 312, 1946–1949 (2006).
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
Latil, M. et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell 20, 191–204.e5 (2017).
Sánchez-Danés, A. et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature 536, 298–303 (2016).
Zhao, X. et al. A transposon screen identifies loss of primary cilia as a mechanism of resistance to SMO inhibitors. Cancer Discov. 7, 1436–1449 (2017).
Whitson, R. J. et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat. Med. 24, 271–281 (2018).
Eberl, M. et al. Tumor architecture and notch signaling modulate drug response in basal cell carcinoma. Cancer Cell 33, 229–243.e4 (2018).
Hoeck, J. D. et al. Stem cell plasticity enables hair regeneration following Lgr5+ cell loss. Nat. Cell Biol. 19, 666–676 (2017).
de Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Shimokawa, M. et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017).
Liu, J. et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl Acad. Sci. USA 110, 20224–20229 (2013).
Northcott, P. A. et al. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012).
Vasioukhin, V., Degenstein, L., Wise, B. & Fuchs, E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA 96, 8551–8556 (1999).
Uhmann, A. et al. The Hedgehog receptor Patched controls lymphoid lineage commitment. Blood 110, 1814–1823 (2007).
Mao, J. et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 (2006).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Aldaz, C. M., Conti, C. J., Gimenez, I. B., Slaga, T. J. & Klein-Szanto, A. J. Cutaneous changes during prolonged application of 12-O-tetradecanoylphorbol-13-acetate on mouse skin and residual effects after cessation of treatment. Cancer Res. 45, 2753–2759 (1985).
Collins, C. A. & Watt, F. M. Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for β-catenin and Notch signalling. Dev. Biol. 324, 55–67 (2008).
Braun, K. M. et al. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development 130, 5241–5255 (2003).
Jensen, K. B., Driskell, R. R. & Watt, F. M. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat. Protoc. 5, 898–911 (2010).
Gonzalez-Roca, E. et al. Accurate expression profiling of very small cell populations. PLoS One 5, e14418 (2010).
Acknowledgements
We thank J.-M. Vanderwinden and M. Martens for help with confocal microscopy. C.B. is an investigator of WELBIO. A.S.-D. and J.-C.L. are supported by fellowships from the FNRS and FRIA, respectively. This work was supported by the FNRS, the Marian Family, the ULB fondation, the foundation Baillet Latour, and a consolidator grant from the European Research Council.
Author contributions
A.S.-D. and C.B. designed the experiments, performed data analysis and wrote the manuscript; A.S.-D. performed most of the biological experiments; J.-C.L, G.L. and V.S. helped with Lgr5 ablation experiments; M.L. performed immunostaining; C.D. performed FACS; E.M.-C., M.S., V.d.M. and J.T. provided patient samples; and A.B. performed GSEA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
C.B. is a consultant at Genentech (San Francisco, USA).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Vismodegib leads to tumour shrinkage and emergence of vismodegib-persistent lesions in mice.
a, Immunostaining for SMOM2, KRT14 and β4-integrin in tail from SmoM2 mice after different durations of vismodegib administration. b, Tumour burden in micrometres (total area occupied by tumours divided by the length of the analysed epidermis) in untreated and vismodegib-treated SmoM2 mice (n = 3 mice analysed per time point and condition). Centre values define the mean. See Source Data for description of the skin length and tumour area analysed per mouse. c, Immunostaining for KRT14 and β4-integrin in ventral skin from Ptch1cKO mice. d, Immunostaining for SMOM2, KRT14 and β4-integrin in tail skin from SmoM2 mice. e, Quantification (mean ± s.e.m.) of lesion type upon vismodegib treatment in SmoM2 mice (n = 3 mice, total number of lesions analysed per time point indicated in parentheses). f, Immunostaining for active caspase-3 (AC3) and SMOM2. g, Percentage of AC3+ tumour cells (mean ± s.e.m.) in untreated and vismodegib-treated SmoM2 mice (n = 30 lesions analysed from 3 mice). Two-sided t-test. h, Immunostaining for Ki67 and SMOM2. i, Percentage of Ki67+ tumour cells (mean ± s.e.m.) in untreated and vismodegib-treated SmoM2 mice (n = 30 lesions analysed from 3 mice). Two-sided t-test. Three independent experiments per condition were analysed showing similar results (a, c, d, f, h). Hoechst nuclear staining in blue; scale bars, 100 μm (c, d), 50 μm (a, f, h). Dashed line delineates basal lamina. Arrows indicate vismodegib-persistent lesions.
Extended Data Fig. 2 Vismodegib-persistent lesions express LGR5 in mice.
a, Immunostaining for LGR5–GFP and β4-integrin at different time points after tamoxifen administration in the Ptch1cKO model. b, In situ hybridization for Lgr5 and Gli1 in untreated and treated tumour cells in SmoM2 mice. c, Percentage (mean ± s.e.m.) of tumour cells (LGR5+ and LGR5–) that express Gli1 in SmoM2 mice (n = 3 mice, total number of cells analysed indicated in parentheses). d, Distribution (mean ± s.e.m.) of the number of Gli1 mRNA dots per tumour cell with and without treatment in SmoM2 mice (n = 104 and 111 total tumour cells from 3 mice per condition and time point). e, Representation of the mouse Smo gene, showing in red the exons (E) in which genetic mutations have been described6,7 (top). Results from the sequencing of exon 7 from vismodegib-persistent lesions obtained by pooling drug-persistent cells from three Krt14CreER;Ptch1cKO;Lgr5DTR-GFP mice, showing absence of genetic mutations in the exon analysed (bottom). See Source Data for the results of the sequencing of exons 3–12. f, Protocol for BrdU and EdU double-labelling studies in Ptch1cKO-induced BCCs followed by vismodegib administration and discontinuation. g, Immunostaining for LGR5–GFP, BrdU and EdU in Ptch1cKO-derived BCCs following 5 days of vismodegib discontinuation. h, Protocol for treatment with vismodegib and retinoic acid (RA) or TPA. i, Immunostaining for LGR5–GFP, Ki67 and β4 in the back skin of Ptch1cKO mice treated with vismodegib and RA or TPA. j, Quantification of LGR5+ tumorigenic lesions per length of skin upon treatment with vismodegib or vismodegib with RA or TPA (n = 3 mice, 3 mm of skin analysed per mouse). Two-sided t-test. k, Immunostaining for LGR5–GFP and LRIG1 in untreated and treated Ptch1cKO mice. l, Immunostaining for LGR5–GFP and LRIG1 in untreated and vismodegib-treated (8 weeks) mice before and after enzymatic and physical separation of epidermis from dermis in Krt14CreER;Ptch1cKO;Lgr5DTR-GFP mice. Note that hair follicles co-expressing LGR5 and LRIG1 and sebaceous cysts remained in the dermal fraction whereas the BCCs were isolated with the epidermal fraction, indicating that normal hair follicles did not significantly contaminate the FACS-isolated tumour cells. m, Cell sorting strategy to isolate LGR5+LRIG1–, LGR5+LRIG1+ and LGR5–LRIG1+ in normal skin and in Ptch1cKO-derived BCCs before and after vismodegib administration. Forward scatter (FSC) and side scatter (SSC) were performed to exclude cell debris and doublets. Living cells were selected by Hoechst dye exclusion. Finally, the different LGR5 and LRIG1 cell populations were isolated by FACS sorting. A, area; W, width. n, Proportion of cells (mean ± s.e.m.) expressing LGR5–GFP and LRIG1 determined by FACS (n = 3 independent experiments per condition). These experiments indicate that LRIG1 can be used to discriminate between LGR5+ cells from the HFSC or lower hair follicle (LGR5+LRIG1–) and BCC cells (LGR5+LRIG1+). Three independent experiments per condition were analysed with similar results (a, k, l). Hoechst nuclear staining in blue; scale bars, 50 μm (a, i, k, l) and 25 μm (b). Dashed line delineates basal lamina separating IFE from the dermis. Dotted line delineates BCC. Arrows indicate vismodegib-persistent lesions.
Extended Data Fig. 3 Characterization of LGR5+LRIG1+ and LGR5–LRIG1+ tumour cells.
a, GSEA showing the enrichment of genes upregulated in the LGR5+LRIG1+ population compared to the LRG5–LRIG1+ population from two independent microarray experiments with the EHFP15 (left) in telogen HFSCs16 (middle) and hair follicle Lgr5-expressing cell signatures17 (right), showing that LGR5-expressing BCC cells express many genes of the embryonic and adult hair follicle signatures. The normalized enrichment score (NES) and P value (one-sided test) were calculated using the GSEA program. b, mRNA expression of genes upregulated in LGR5+LRIG1+ tumour cells compared to LGR5–LRIG1+ tumour cells in untreated conditions (n = 2 independent microarray experiments). c, Immunostaining for LGR5–GFP with LEF1, LHX2, CUX1, TBX1 and ALCAM in untreated Ptch1cKO-derived BCCs. d, Venn diagram showing the similarities and differences between genes that were upregulated more than twofold from two independent microarray experiments in LGR5–LRIG1+ versus LGR5+LRIG1+ cells compared to IFE16 and LRIG113 signatures. P value calculated using the hypergeometric test for each intersection of two subsets of genes with phyper function in R software. The high overlap indicates that LGR5–LRIG1+ cells expressed IFE and infundibulum differentiation markers. e, mRNA expression of genes upregulated in LGR5–LRIG1+ tumour cells compared to LGR5+LRIG1+ cells in untreated conditions (n = 2 independent microarray experiments). f, PCR analysis of the recombination of the floxed Ptch1 alleles in control samples and in FACS-isolated tumour-derived LGR5+LRIG1+ and LGR5–LRIG1+ populations from Ptch1cKO-induced BCCs. Two technical replicates were analysed for each sample with similar results. g, Immunostaining for LGR5–GFP, LRIG1 and Ki67 in Ptch1cKO-derived BCCs shows higher proliferation rate in LGR5+LRIG1+ than in LGR5–LRIG1+ tumour cells. Three independent experiments per condition were analysed with similar results (c, g). Hoechst nuclear staining in blue; scale bars, 50 μm.
Extended Data Fig. 4 Transplantation of LGR5+LRIG1+ Ptch1;Trp53 double conditional knockout BCC cells leads to the formation of BCC-like structures.
a, Table summarizing the number of grafted mice that presented KRT14+ BCC-like structures upon transplantation of FACS-isolated LGR5+LRIG1+ and LGR5–LRIG1+ cells from BCCs arising in Krt14CreER;Ptch1cKO;Lgr5DTR–GFP and Krt14CreER;Ptch1cKO;Trp53cKO;Lgr5DTR–GFP mice. b, c, Immunostaining for LGR5–GFP, KRT14 and LRIG1 (b) and for LGR5–GFP and KRT10 (c) in the BCC-like structures obtained upon transplantation of LGR5+LRIG1+ cells from Ptch1;Trp53 double conditional knockout BCCs in the dorsal fat pads of NOD/SCID mice. Three independent experiments per condition were analysed with similar results (b, c). Hoechst nuclear staining in blue; scale bars, 50 μm.
Extended Data Fig. 5 Vismodegib-persistent lesions do not show decreased primary cilia numbers or nuclear localization of MKL1.
a, b, Immunostaining for ARL13B and LGR5–GFP in Ptch1cKO model (a) and for ARL13B and SMOM2 in SmoM2 model (b) in untreated and vismodegib-treated lesions. c, d, Immunostaining for MKL1 and LGR5–GFP in vismodegib-persistent lesions in Ptch1cKO mice (c) and for MKL1 and SMOM2 in vismodegib-persistent lesions in SmoM2 mice (d) treated for 8 weeks with vismodegib. White boxes are expanded on right. Three independent experiments per condition were analysed with similar results. Scale bars, 25 μm.
Extended Data Fig. 6 Vismodegib promotes BCC differentiation.
a, Immunostaining for LGR5–GFP, LHX2 and ALCAM in untreated and vismodegib-treated Ptch1cKO-derived BCCs. b, GSEA showing enrichment of genes upregulated in LGR5+LRIG1+ vismodegib-treated tumours compared to untreated BCCs with IFE16 and LRIG113 signatures, showing that vismodegib treatment promotes the expression of the IFE and infundibulum signatures. The normalized enrichment score (NES) and P value (one-sided test) were calculated using the GSEA program. c, Oil Red O and haematoxylin and eosin staining in ventral skin of untreated and vismodegib-treated Ptch1cKO mice. Arrows indicate areas of sebaceous differentiation. d, Immunostaining for LGR5–GFP and adipophilin in untreated and vismodegib-treated Ptch1cKO-derived BCCs. Arrows indicate areas of sebaceous differentiation. e, In situ hybridization for Lgr5 and Defb6 in untreated and vismodegib-treated Ptch1cKO-derived BCCs. f, Immunostaining for KRT1 and SMOM2 in untreated and vismodegib-treated SmoM2 mice. Three independent experiments per condition were analysed with similar results (a, c–f). Hoechst nuclear staining in blue; scale bars, 50 μm.
Extended Data Fig. 7 Vismodegib promotes differentiation of SMOM2-expressing cells during BCC initiation and in Ptch1cKO tumour cells.
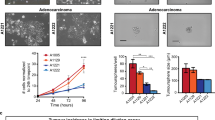
a, Protocol for tumour induction and timing of vismodegib administration to Krt14CreER;RosaSmoM2 mice. b, Quantification of surviving SMOM2 clones in the interscale (tail epidermis) in untreated mice and after different durations of vismodegib treatment (n = 3 mice per time point and condition). Centre values show mean. See Source Data for description of total number of clones counted per time point and condition. c, Immunostaining for KRT31 and SMOM2 in whole-mount tail skin (left) and orthogonal views of the clones highlighted in the left panel stained for β4-integrin and SMOM2 (right). d, Quantification (mean ± s.e.m.) of the type of SMOM2-expressing clones after different durations of vismodegib treatment (n = 3 or 4 mice as indicated in the graph, total number of lesions quantified indicated in parentheses). e, f, Immunostaining for KRT10 and SMOM2 (e) and for LHX2 and SMOM2 (f) in untreated and vismodegib-treated mice. Three independent experiments per condition were analysed with similar results. g, mRNA expression of genes upregulated in the LGR5+LRIG1+CD71+ population compared to the LGR5+LRIG1+CD71– population obtained by quantitative PCR (n = 3 mice). Bars represent the average fold change over LGR5+LRIG1+CD71– cells and error bars the s.e.m. h, Immunostaining for LGR5–GFP, BrdU and KRT10 in mice that received three injections of BrdU followed by two weeks of vismodegib administration. Three independent experiments per condition were analysed with similar results. Hoechst nuclear staining in blue; scale bars, 100 μm (c) and 50 μm (e, f, h).
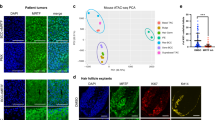
Extended Data Fig. 8 LGR5 expression in vismodegib-persistent lesions in human BCCs.
a, Tables summarizing the BCC and treatment characteristics in the patients analysed. b, Immunohistochemistry for KRT14 in biopsies before, after and during vismodegib treatment. c, In situ hybridization for LGR5 and GLI1 in biopsies from patients before, during and after vismodegib treatment. d, Percentage (mean ± s.e.m.) of tumour cells (LGR5+ and LGR5–) that express GLI1 in biopsies from patients, during or after vismodegib treatment (n = 3 samples from different patients (Patients 1–3) or body locations (Patient 4), total number of cells analysed indicated in parentheses). e, f, Immunohistochemistry for Ki67 (e) and KRT10 (f) in biopsies before, during and after vismodegib treatment. Hoechst nuclear staining in blue; scale bars, 25 μm.
Extended Data Fig. 9 Lgr5 lineage ablation leads to BCC shrinkage and elimination of vismodegib-persistent lesions.
a, Protocol for tamoxifen and diphtheria toxin (DT) administration. b, c, Immunostaining for KRT14 and LGR5–GFP in the Ptch1cKO model (b) and for KRT14 and SMOM2 in the SmoM2 model (c) after different durations of DT administration. d, Quantification of tumour burden in untreated mice and after DT administration (n = 3 mice per time point and condition). Centre values define the mean. See Source Data for description of the skin length and tumour area analysed per mouse. e, Number of LGR5–GFP+ tumour cells in untreated conditions and following DT administration (n = 3 mice per time point and condition, 1 mm of skin analysed per mouse). Centre values define the mean. f, Quantification of tumour burden (SMOM2-expressing cells) in untreated conditions and following DT treatment (n = 3 mice per time point and condition). Centre values define the mean. See Source Data for description of the skin length and tumour area analysed per mouse. g, Immunostaining for active caspase-3 (AC3) and LGR5–GFP (top) and for active caspase-3 and SMOM2 (bottom) after five administrations of DT. Three independent experiments per condition were analysed with similar results. h, Experimental strategy for combination of vismodegib treatment and Lgr5 ablation in Krt14CreER;Ptch1cKO;Lgr5DTR–GFP mice. i, Immunostaining for LGR5–GFP and KRT14 in Krt14CreER;Ptch1cKO;Lgr5DTR–GFP mice upon treatment, Lgr5 ablation and discontinuation. j, Immunostaining for active caspase-3 and LGR5–GFP following administration of vismodegib and DT. k, Quantification of the number of LGR5–GFP+ cells (mean ± s.e.m.) in the different experimental conditions upon treatment and discontinuation (n = 3 mice, 3 mm of skin analysed per mouse). Two-sided t-test. l, Quantification of the number of LGR5+ lesions (mean ± s.e.m.) per length of epidermis (mm) in mice treated with vismodegib and upon discontinuation of treatment with vismodegib and DT (n = 3 mice, 3 mm of skin analysed per mouse). Two-sided t-test. Hoechst nuclear staining in blue; scale bars, 50 μm. Dashed line delineates basal lamina separating IFE from the dermis. Dotted line delineates BCC. Arrows indicate tumorigenic lesions in b, c and indicate vismodegib-persistent lesions in i.
Extended Data Fig. 10 Wnt signalling is active in vismodegib-persistent lesions in mouse and human BCCs.
a, ISH for Lgr5 and Axin2 in untreated and vismodegib-treated lesions from Ptch1cKO and SmoM2 mice. b, ISH for LGR5 and AXIN2 in biopsies from patients before, during and after vismodegib treatment. c, Immunostaining for LEF1 and LGR5–GFP in Ptch1cKO-derived tumorigenic lesion following treatment with vismodegib and LGK-974. d, Protocol used for LGK-974 treatment in Ptch1cKO mice. e, Immunostaining for LGR5–GFP and KRT14 in BCC treated with LGK-974 for 9 days from the Ptch1cKO model. f, Quantification of the tumour burden (mean ± s.e.m.) in mice treated with LGK-974 for 9 days or untreated (n = 3 mice). See Source Data for description of skin length and tumour area analysed per mouse. Two-sided t-test. Hoechst nuclear staining in blue; scale bars, 25 μm. Dashed line delineates basal lamina separating IFE from the dermis. Dotted line delineates BCC. Three independent experiments per condition were analysed with similar results (a, c, e) and two technical replicates were performed for each sample with similar results (b).
Extended Data Fig. 11 Model.
Vismodegib administration promotes tumour cell differentiation leading to BCC regression. However, upon vismodegib treatment a small proportion of LGR5+ BCC cells persists, forming vismodegib-tolerant lesions that are slow cycling and characterized by Wnt signalling activation. Discontinuation of vismodegib treatment results in proliferation of LGR5-persistent lesions that lead to BCC relapse. Vismodegib treatment in combination with Lgr5 lineage ablation or Wnt signalling inhibition results in eradication of BCCs.
Supplementary information
Rights and permissions
About this article
Cite this article
Sánchez-Danés, A., Larsimont, JC., Liagre, M. et al. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature 562, 434–438 (2018). https://doi.org/10.1038/s41586-018-0603-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0603-3
Keywords
This article is cited by
-
Cancer cell plasticity, stem cell factors, and therapy resistance: how are they linked?
Cancer and Metastasis Reviews (2024)
-
Cancer cell plasticity: from cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance
Cancer and Metastasis Reviews (2024)
-
Modeling stress-induced responses: plasticity in continuous state space and gradual clonal evolution
Theory in Biosciences (2024)
-
Rational combinations of targeted cancer therapies: background, advances and challenges
Nature Reviews Drug Discovery (2023)
-
Cancer cell plasticity during tumor progression, metastasis and response to therapy
Nature Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.