Abstract
Histone H3 lysine 9 methylation (H3K9me) mediates heterochromatic gene silencing and is important for genome stability and the regulation of gene expression1,2,3,4. The establishment and epigenetic maintenance of heterochromatin involve the recruitment of H3K9 methyltransferases to specific sites on DNA, followed by the recognition of pre-existing H3K9me by the methyltransferase and methylation of proximal histone H35,6,7,8,9,10,11. This positive feedback loop must be tightly regulated to prevent deleterious epigenetic gene silencing. Extrinsic anti-silencing mechanisms involving histone demethylation or boundary elements help to limit the spread of inappropriate H3K9me12,13,14,15. However, how H3K9 methyltransferase activity is locally restricted or prevented from initiating random H3K9me—which would lead to aberrant gene silencing and epigenetic instability—is not fully understood. Here we reveal an autoinhibited conformation in the conserved H3K9 methyltransferase Clr4 (also known as Suv39h) of the fission yeast Schizosaccharomyces pombe that has a critical role in preventing aberrant heterochromatin formation. Biochemical and X-ray crystallographic data show that an internal loop in Clr4 inhibits the catalytic activity of this enzyme by blocking the histone H3K9 substrate-binding pocket, and that automethylation of specific lysines in this loop promotes a conformational switch that enhances the H3K9me activity of Clr4. Mutations that are predicted to disrupt this regulation lead to aberrant H3K9me, loss of heterochromatin domains and inhibition of growth, demonstrating the importance of the intrinsic inhibition and auto-activation of Clr4 in regulating the deposition of H3K9me and in preventing epigenetic instability. Conservation of the Clr4 autoregulatory loop in other H3K9 methyltransferases and the automethylation of a corresponding lysine in the human SUV39H2 homologue16 suggest that the mechanism described here is broadly conserved.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Moazed, D. Mechanisms for the inheritance of chromatin states. Cell 146, 510–518 (2011).
Zhang, K., Mosch, K., Fischle, W. & Grewal, S. I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388 (2008).
Al-Sady, B., Madhani, H. D. & Narlikar, G. J. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol. Cell 51, 80–91 (2013).
Ragunathan, K., Jih, G. & Moazed, D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015).
Audergon, P. N. et al. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015).
Wang, X. & Moazed, D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356, 88–91 (2017).
Jih, G. et al. Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature 547, 463–467 (2017).
Thon, G., Bjerling, P., Bünner, C. M. & Verhein-Hansen, J. Expression-state boundaries in the mating-type region of fission yeast. Genetics 161, 611–622 (2002).
Trewick, S. C., Minc, E., Antonelli, R., Urano, T. & Allshire, R. C. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 26, 4670–4682 (2007).
Wang, J. et al. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev. 27, 1886–1902 (2013).
Zofall, M. & Grewal, S. I. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692 (2006).
Piao, L. et al. Automethylation of SUV39H2, an oncogenic histone lysine methyltransferase, regulates its binding affinity to substrate proteins. Oncotarget 7, 22846–22856 (2016).
Min, J., Zhang, X., Cheng, X., Grewal, S. I. & Xu, R. M. Structure of the SET domain histone lysine methyltransferase Clr4. Nat. Struct. Biol. 9, 828–832 (2002).
Zhang, X. et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12, 177–185 (2003).
Li, Y. et al. Structural basis for activity regulation of MLL family methyltransferases. Nature 530, 447–452 (2016).
Moritz, L. E. & Trievel, R. C. Structure, mechanism, and regulation of polycomb repressive complex 2. J. Biol. Chem. https://doi.org/10.1074/jbc.R117.800367 (2017).
Wang, J., Reddy, B. D. & Jia, S. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife 4, (2015).
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Wu, H. et al. Structural biology of human H3K9 methyltransferases. PLoS ONE 5, e8570 (2010).
Southall, S. M., Wong, P. S., Odho, Z., Roe, S. M. & Wilson, J. R. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol. Cell 33, 181–191 (2009).
Bintu, L. et al. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724 (2016).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Morgan, D. O. Principles of CDK regulation. Nature 374, 131–134 (1995).
Shan, C. M. et al. A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading. eLife 5, e17903 (2016).
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Iglesias, N. et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24, 1927–1938 (2010).
Ting, L., Rad, R., Gygi, S. P. & Haas, W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
Paulo, J. A., O’Connell, J. D. & Gygi, S. P. A triple knockout (TKO) proteomics standard for diagnosing ion interference in isobaric labeling experiments. J. Am. Soc. Mass Spectrom. 27, 1620–1625 (2016).
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010).
Beausoleil, S. A., Villén, J., Gerber, S. A., Rush, J. & Gygi, S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006).
Paulo, J. A., O’Connell, J. D., Gaun, A. & Gygi, S. P. Proteome-wide quantitative multiplexed profiling of protein expression: carbon-source dependency in Saccharomyces cerevisiae. Mol. Biol. Cell 26, 4063–4074 (2015).
Wong, K. H., Jin, Y. & Moqtaderi, Z. Multiplex Illumina sequencing using DNA barcoding. Curr. Protoc. Mol. Biol. 101, 7.11.1–7.11.11 (2013).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Acknowledgements
We are grateful for assistance from the staff at NE-CAT at Argonne National Laboratory, the SBGrid consortium at Harvard Medical School, and S. Jenni for useful discussions, H. Yoon for help with the scintillation counter, Z. Moqtaderi and R. Yu for Python scripts, G. Shipkovenska, A. Tatarakis, X. Wang, A. Yuan and H. Zhou for comments on the manuscript, and members of the Moazed laboratory for discussion. This work used NE-CAT beamlines (GM103403), a Pilatus detector (RR029205), an Eiger detector (OD021527) and APS Synchrotron source (DE-AC02-06CH11357). This work was supported by an EMBO long-term fellowship and a Swiss National Science Foundation postdoctoral fellowship (N.I.), K01 DK098285 (J.A.P.), NIH P50 GM107618 (M.K., S.P.G.) and NIH RO1 GM072805 (D.M.). D.M. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
N.I., M.A.C. and D.M. designed experiments. N.I. prepared recombinant proteins for methyltransferase assays, performed methylation, mutagenesis, strain construction, ChIP–seq, western blot, silencing and growth assays. G.J. performed strain construction and ChIP–seq for Extended Data Fig. 8a–d. N.I. analysed all other sequencing libraries. M.A.C. prepared and crystallized Clr4192–490 proteins, collected X-ray diffraction data, solved structures and performed molecular modelling and structural analysis. J.A.P., N.S. and M.K. performed mass spectrometry. S.P.G. and D.M. supervised research. N.I., M.A.C. and D.M. wrote and edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Analysis of Clr4 automethylation.
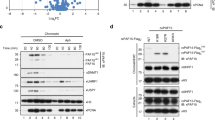
a, Outline of the in vitro methyltransferase activity assays. b, In vitro methyltransferase assay showing that Clr4dead (Clr4(Y451N)) has no methyltransferase activity towards itself or histone H31–20 peptide and that wild-type Clr4 with or without an N-terminal GST tag displays similar methyltransferase activities. Experiment repeated twice with similar results. c, Outline of the in vitro methyltransferase assay with unmethylated or automethylated Clr4. d, Time course of methyltransferase activity of GST–Clr4 towards H31–21 peptide substrate (10.8 μM) after a 1-h incubation of GST–Clr4 with 512 μM cold SAM (+) or without SAM (−), showing that the automethylation of Clr4 increases its methyltransferase activity on the H31–21 substrate. Incorporation of 3H-SAM on H31–21 peptide was assessed by scintillation counting. Data are mean ± s.d. from three biological replicates. e, Quantitative mass spectrometry analysis of E. coli-purified GST–Clr4 wild type and GST–Clr4dead, showing that Clr4 K455 is the sole target of automethylation detected in our experiments and that the remaining sites were methylated by E. coli enzymes. Data are mean ± s.e.m. from three (wild type, orange circles) or six (Clr4dead, grey circles) biological replicates. Data are presented as a ratio (upper axis) of values for each peptide (lower axis) of Clr4 wild type versus GST–Clr4dead containing the indicated methylated lysine. f, g, In vitro methyltransferase assays showing that Clr4 K127 is not a major target of automethylation, as substitution of K127 with arginine (K127R) (f) or deletion of a part of the hinge and the chromodomain (Clr4192–490) that contains K127 (g) did not affect the level of Clr4 automethylation. Experiment performed once (f) and more than three times with similar results (g). See Supplementary Fig. 1 for the uncropped gels.
Extended Data Fig. 2 Clr4 K455 is a target of automethylation.
a–c, LC–MS/MS analysis showing monomethylation (a), dimethylation (b) and trimethylation (c) of Clr4 at K455. The peptide sequence (amino acids 440–464) with theoretical masses of MS/MS fragments (top) and the MS/MS spectrum corresponding to the monomethylated (a), dimethylated (b) and trimethylated (c) Clr4 K455 are shown (bottom). Experiment repeated more than four times (with either LysC or trypsin digestion) with similar results. d, Kinetics of in vitro GST–Clr4 automethylation followed by quantitative mass spectrometry analysis showing an increase in automethylation of Clr4 K455 at the indicated time points. In the case of K455me1 and me2, two different-sized peptides were quantified and are indicated by #1 and #2 in each case. The lower levels of K455me3, relative to K455me1 and K455me2, are likely to reflect faster rates of mono- and dimethylation relative to trimethylation. The small increase in K455me3 without a corresponding decrease in K455me1 or K455me2 suggests that automethylation did not reach saturation in our reactions. Data are mean ± s.e.m. from six biological replicates.
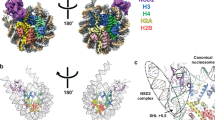
Extended Data Fig. 3 Comparison of Clr4 structure with other SUV39H methyltransferases.
a, Alignment of the autoinhibited Clr4192–490 (yellow) and the previously determined Clr4192–490 structure that lacks post-SET domain and cofactor (grey) (PDB ID: 1MVH). b, Close-up view of S. pombe Clr4192–490 autoinhibited conformation (surface) showing that K455 (red stick) in the autoregulatory loop (red) is located within the active site pocket. c, Alignment of autoinhibited Clr4192–490 (yellow) and DIM-5 (grey) in complex with histone H3 peptide (magenta) and SAH (PDB ID: 1PEG). d, e, Schematic of residues that engage in interactions between Clr4 (grey, yellow and cyan) and Clr4 autoregulatory loop (red) (d) and DIM-5 (grey) and histone H3 (magenta) (e). Hydrogen bonds and salt bridge interactions are shown as dashed lines. Colour assignments are as in Fig. 1d; residues in the SET insertion domain, SET and post-SET domain are shown in yellow, grey and cyan, respectively. SAH is shown in dark blue. f, Alignment of the catalytic pockets of Clr4192–490 and DIM-5 (PDB ID: 1PEG) showing that Clr4 K455 (red) and K9 of the histone H3 substrate (magenta) occupy similar positions. g–i, Close-up view of the autoinhibited conformation of Clr4 active site (g, as in Fig. 1h, shown here for comparison) and modelling the K455A (h) and K455R (i) mutants in the active site. Possible bonding interactions are shown as dashed lines.
Extended Data Fig. 4 Conformational change in Clr4 induced by automethylation.
a, Surface representation of the active-site area of Clr4 in autoinhibited Clr4 (left) and automethylated Clr4 (right), showing the opening of the catalytic pocket induced by automethylation; the methyl group of SAM is visible in the active-site tunnel. b, Simulated annealing composite omit map for the autoregulatory-loop-occupied active site of Clr4 contoured at 1.0σ. The protein is coloured as in Fig. 1d and the electron density is shown in blue. c, The simulated annealing composite omit map of the same region in the automethylated Clr4 crystal structure, contoured at 1.0σ, lacks density for autoregulatory loop residues. d, e, Polder omit map for the bound cofactors of the autoinhibited (d) and automethylated (e) crystal structures contoured at 3.5σ.
Extended Data Fig. 5 Analysis of Clr4 autoregulatory-loop-mutant proteins and their effect on histone H3K9me2 or me3 in vitro and in vivo.
a, In vitro methyltransferase assay showing that the addition of the K472R mutation reduced both the automethylation and methyltransferase activity of the hyperactive Clr4(K455A) protein towards the H31–20 peptide. Exposure time is indicated. Experiment repeated twice with similar results. b, Methyltransferase activity of the Clr4(K472R) mutant is not stimulated after automethylation (red +). Exposure times are indicated. Experiment performed once. c, Western blot of N-terminally 3× Flag-tagged Clr4 showing that mutations of the autoregulatory loop (K455 and K472 to alanine or arginine) do not affect Clr4 protein stability (top). The same blot stained with Ponceau dye is shown as a loading control (bottom). Experiment performed once. d, Diagram of Clr4 with K455 and K472, with arginine, alanine or tryptophan (for K455 only) substitutions and their expected effect on the methyltransferase activity of Clr4. e, H3K9me2 ChIP–seq reads mapped to different heterochromatin regions in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data), and highlight the increased or decreased spreading of H3K9me2 in the Clr4 K455 and K472 mutants. Top, chromosome coordinates. # indicates an independent clone. Sequencing performed twice with similar results with two independent clones. f, As in e, but showing H3K9me3 ChIP–seq data of tel1L and tel2L (left) and an expanded view of H3K9me3 ChIP–seq (right). See legend of Fig. 3 for abbreviations. Sequencing repeated twice with two to three independent clones with similar results. See Supplementary Fig. 1 for the uncropped gels.
Extended Data Fig. 6 Hyperactive Clr4 mutants promote epigenetic instability.
a, Left, map of the right arm of centromere 1 (cen1) with the insertion of the ura4+ reporter gene at the pericentromeric otr1R repeat (otr1R::ura4+). Right, otr1R::ura4+ silencing assay in the indicated genotypes, showing clone-to-clone variation of ura4+ silencing in hyperactive clr4 mutants. N/S, nonselective medium; -Ura, medium without uracil; +FOA, medium containing 5-FOA. # indicates an independent clone. Experiment performed twice with similar results with two to three independent clones, as shown. b, H3K9me3 ChIP–seq reads mapped to different heterochromatin regions (left) or euchromatic genes (right) in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data) and highlight increased or decreased spreading of H3K9me2 or me3 in Clr4 K455 mutants. S. pombe h+ cells used for the ChIP–seq experiments have a duplicated mat locus. Top, chromosome coordinates. See legend of Fig. 3 for abbreviations. Sequencing performed once with two to three independent clones as shown.
Extended Data Fig. 7 Disruption of Clr4 autoinhibition results in growth defects, inappropriate H3K9me spreading and formation of new H3K9me domains.
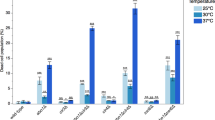
a, Growth assays showing that cells that express the indicated hyperactive Clr4 mutants have growth defects. # indicates an independent clone. Data are mean ± s.d. from three biological replicates. b, Silencing assay of mat2P::ade6+ on low-adenine medium with one (1×) or two (2×) copies of clr4+ or mutant clr4 genes. See main text for details. Experiment performed twice with at least two independent clones with similar results. c, d, H3K9me3 (c) or H3K9me2 (d) ChIP–seq reads mapped to different heterochromatin regions (left) or euchromatic genes (right) in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data). Top, chromosome coordinates. See legend of Fig. 3 for abbreviations. Sequencing performed once with two independent clones, as shown. e, Growth assays showing that the addition of a second copy (2×) of the clr4 gene that encodes hyperactive Clr4 mutants exacerbates growth defects. # indicates an independent clone. Data are mean ± s.d. from three biological replicates.
Extended Data Fig. 8 Deletion of the Epe1 anti-silencing pathway leads to epigenetic instability.
a, b, H3K9me2 (top, blue) and H3K9me3 (bottom, red) ChIP–seq reads mapped to different heterochromatin regions (a) or euchromatic genes (b) in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data). Top, chromosome coordinates. mit1+ encodes a Clr3 HDAC-associated factor required for efficient heterochromatic silencing; erm1+ encodes an endoplasmic reticulum peptidase; and utp22+ encodes a ribosomal RNA processing factor. Note that epe1+ #1, epe1Δ #4 and epe1Δ #5 clones were constructed in the SPY137 background (which has an insertion of ura4+ at the pericentromeric region of chromosome 1) and epe1+ #2 and epe1Δ #1–epe1Δ #3 clones were constructed in the SPY3 background (which has an insertion of ade6+ at the mat locus, see Supplementary Table 3 for detailed genotypes). Sequencing performed once with the indicated independent clones (#) for each genotype. c, d, H3K9me2 ChIP–seq in the indicated genotypes showing clone-to-clone variation in the appearance of H3K9me2 peaks throughout the S. pombe genome (coloured arrowheads). H3K9me2 ChIP–seq reads mapped to all three S. pombe chromosomes in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data). Top, chromosome coordinates. Black arrowhead with asterisk indicates the location of the ura4+ gene. Strains were constructed by crossing cells with and without a ura4+ insertion at pericentromeric repeats. Therefore, not all progeny of the cross show H3K9me2 mapping to ura4+ (highlighted with asterisk on the left arm of chromosome 3). See Supplementary Table 2 for a list of H3K9me peaks. Sequencing performed once with the indicated independent clones (#) for each genotype. e, Silencing assay of mat2P::ade6+ on low-adenine medium, showing that the combination of epe1+ deletion (∆) and a hyperactive clr4 mutant results in clone-to-clone variegation of the ade6+ silencing defect. Experiment performed once with five independent clones as shown. f, H3K9me2 ChIP–seq reads mapped to different heterochromatin regions (left) or euchromatic genes (right) in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data). Top, chromosome coordinates. See legend of Fig. 3 for abbreviations. Sequencing performed once with three to four independent clones. # indicates an independent clone.
Extended Data Fig. 9 Clr4 autoregulation prevents illegitimate heterochromatin formation mediated by the Clr4 read–write positive feedback mechanism.
a, b, H3K9me3 ChIP–seq reads mapped to different heterochromatin regions (a) or euchromatic genes (b) in the indicated genotypes are presented as reads per million (number in brackets in the first row of each set of ChIP–seq data). Top, chromosome coordinates. See legend of Fig. 3 for abbreviations. Sequencing performed once with the indicated independent clones (#) for each genotype. c, Schematic showing Clr4 chromodomain (CD)-dependent spreading and maintenance of histone H3K9me. The CD-dependent positive feedback loop is critical even with hyperactive Clr4.
Extended Data Fig. 10 Clr4 automethylation-dependent heterochromatin regulation and evidence for its evolutionary conservation.
a, Schematic of the role of lysine automethylation in the autoregulatory loop, in preventing illegitimate heterochromatin formation and epigenetic instability (left). Intrinsic regulation of Clr4 by automethylation (of K455 and probably of K472) acts in parallel with other anti-silencing mechanisms that involve that regulation of Clr4 levels and H3K9 demethylation (red boxes). See main text for additional discussion. b, Top, diagram illustrating the domain organization of the S. pombe Clr4 protein and the location of Clr4 K455 (red arrowhead) and K472 (orange arrowhead). Bottom, sequence alignment of the Clr4 autoregulatory loop that contains K455 (red) and K472 (orange) in the indicated methyltransferases. SUV39H2 K375 and SUV39H2 K392 are indicated in underlined bold. H. sapiens, Homo sapiens; M. musculus, Mus musculus; N. crassa, Neurospora crassa. c, Overlay of S. pombe Clr4 in grey (this study, PDB ID: 6BOX) and human SUV39H2 in yellow (PDB ID: 2R3A), showing that S. pombe Clr4 K455 (shown as red stick) and human SUV39H2 K375 (shown as yellow stick) occupy partially overlapping positions inside the catalytic pocket.
Supplementary information
Supplementary Figure 1
The uncropped gels for the panels used in Fig. 1a, b, c, e, f, j, Fig. 3a, b, Extended Data Fig. 1b, f, g, and Extended Data Fig. 5a, b, c.
Supplementary Table 1
Genome-wide distribution of changes in H3K9me ChIP–seq peaks in the indicated strains containing one or two copies of wild-type or mutant clr4 is summarized.
Supplementary Table 2
Genome-wide distribution of changes in H3K9me ChIP–seq peaks in the indicated epe1+ and epe1Δ cells is summarized.
Supplementary Table 3
List of fission yeast S. pombe strains used in this study.
Supplementary Table 4
List of plasmids used in this study.
Supplementary Table 5
Quantitative mass spectrometry analysis of Clr4-K455 methylation states in Clr4 crystals.
Supplementary Table 6
Crystallographic data collection and refinement statistics.
Rights and permissions
About this article
Cite this article
Iglesias, N., Currie, M.A., Jih, G. et al. Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature 560, 504–508 (2018). https://doi.org/10.1038/s41586-018-0398-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0398-2
This article is cited by
-
Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
Nature Reviews Molecular Cell Biology (2022)
-
Arsenic 3 methyltransferase (AS3MT) automethylates on cysteine residues in vitro
Archives of Toxicology (2022)
-
Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation
Nature Genetics (2020)
-
Unprogrammed epigenetic variation mediated by stochastic formation of ectopic heterochromatin
Current Genetics (2020)
-
Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription
Communications Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.