Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) controls cell growth and metabolism in response to nutrients, energy levels, and growth factors. It contains the atypical kinase mTOR and the RAPTOR subunit that binds to the Tor signalling sequence (TOS) motif of substrates and regulators. mTORC1 is activated by the small GTPase RHEB (Ras homologue enriched in brain) and inhibited by PRAS40. Here we present the 3.0 ångström cryo-electron microscopy structure of mTORC1 and the 3.4 ångström structure of activated RHEB–mTORC1. RHEB binds to mTOR distally from the kinase active site, yet causes a global conformational change that allosterically realigns active-site residues, accelerating catalysis. Cancer-associated hyperactivating mutations map to structural elements that maintain the inactive state, and we provide biochemical evidence that they mimic RHEB relieving auto-inhibition. We also present crystal structures of RAPTOR–TOS motif complexes that define the determinants of TOS recognition, of an mTOR FKBP12–rapamycin-binding (FRB) domain–substrate complex that establishes a second substrate-recruitment mechanism, and of a truncated mTOR–PRAS40 complex that reveals PRAS40 inhibits both substrate-recruitment sites. These findings help explain how mTORC1 selects its substrates, how its kinase activity is controlled, and how it is activated by cancer-associated mutations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ben-Sahra, I. & Manning, B. D. mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 45, 72–82 (2017)
Bar-Peled, L. & Sabatini, D. M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24, 400–406 (2014)
Dibble, C. C. & Cantley, L. C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25, 545–555 (2015)
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012)
Grabiner, B. C. et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 4, 554–563 (2014)
Keith, C. T. & Schreiber, S. L. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270, 50–51 (1995)
Kim, D. H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002)
Hara, K. et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 (2002)
Kim, D. H. et al. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11, 895–904 (2003)
Aylett, C. H. et al. Architecture of human mTOR complex 1. Science 351, 48–52 (2016)
Saucedo, L. J. et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5, 566–571 (2003)
Stocker, H. et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5, 559–566 (2003)
Sancak, Y. et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 (2007)
Vander Haar, E., Lee, S. I., Bandhakavi, S., Griffin, T. J. & Kim, D. H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 (2007)
Wang, L., Harris, T. E., Roth, R. A. & Lawrence, J. C. Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 282, 20036–20044 (2007)
Oshiro, N. et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 282, 20329–20339 (2007)
Burnett, P. E ., Barrow, R. K ., Cohen, N. A ., Snyder, S. H. & Sabatini, D. M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl Acad. Sci. USA 95, 1432–1437 (1998)
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009)
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011)
Kang, S. A. et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566 (2013)
Schalm, S. S., Fingar, D. C., Sabatini, D. M. & Blenis, J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 (2003)
Nojima, H . et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 (2003)
Yang, H. et al. mTOR kinase structure, mechanism and regulation. Nature 497, 217–223 (2013)
Yang, H. et al. 4.4 Å resolution cryo-EM structure of human mTOR complex 1. Protein Cell 7, 878–887 (2016)
Choi, J., Chen, J., Schreiber, S. L. & Clardy, J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242 (1996)
Yu, Y. et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 (2011)
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012)
Michels, A. A. MAF1: a new target of mTORC1. Biochem. Soc. Trans. 39, 487–491 (2011)
Peterson, T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011)
Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716 (1999)
Ginalski, K., Zhang, H. & Grishin, N. V. Raptor protein contains a caspase-like domain. Trends Biochem. Sci. 29, 522–524 (2004)
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012)
Scheres, S. H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016)
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015)
Baretic´, D., Berndt, A., Ohashi, Y., Johnson, C. M. & Williams, R. L. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat. Commun. 7, 11016 (2016)
Yu, Y. et al. Structural basis for the unique biological function of small GTPase RHEB. J. Biol. Chem. 280, 17093–17100 (2005)
Klink, T. A., Kleman-Leyer, K. M., Kopp, A., Westermeyer, T. A. & Lowery, R. G. Evaluating PI3 kinase isoforms using Transcreener ADP assays. J. Biomol. Screen. 13, 476–485 (2008)
Wagle, N. et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 4, 546–553 (2014)
Ohne, Y. et al. Isolation of hyperactive mutants of mammalian target of rapamycin. J. Biol. Chem. 283, 31861–31870 (2008)
Urano, J. et al. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl Acad. Sci. USA 104, 3514–3519 (2007)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016)
Scheres, S. H. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3, e03665 (2014)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014)
Schägger, H. Tricine–SDS-PAGE. Nat. Protocols 1, 16–22 (2006)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Acknowledgements
We thank the staff of the Advanced Photon Source, the New York Structural Biology Center Simons Electron Microscopy Center, the Howard Hughes Medical Institute Cryo-EM facility, the Memorial Sloan Kettering Cancer Center Cryo-EM facility, and Subangstrom for help with data collection. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health grant CA008748.
Author information
Authors and Affiliations
Contributions
H.Y., B.L., and N.P.P. designed the experiments, determined the mTOR structures, and wrote the manuscript. N.P.P. and X.J. determined the Raptor crystal structures and biochemical constants. H.Y., H.J.Y., M.M., A.Y., and A.D. performed the mTOR enzyme assays and biochemical experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Barford, N. MacDonald and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
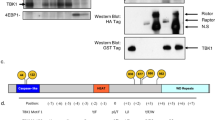
Extended Data Figure 1 Substrate recruitment by the FRB domain.
a, Deletion mapping of S6K1 FRB-binding motif polypeptides (2 μM) using phosphorylation by the mTORΔN–mLST8 (30 nM) as the assay, extending previous findings23. Truncation up to 7 residues N-terminal to the Thr389 phosphorylation site (indicated by asterisk) has minimal effect, whereas C-terminal truncations starting 15 residues from Thr389 successively reduce phosphorylation. The polypeptides were produced as described in Methods for the S6K1367–404 peptide. Column graph shows velocity divided by enzyme concentration from the quantification of the 32P autoradiogram. Columns show means and markers show values from independent experiments (n = 6 for 382–410, 367–392, 367–398, and 367–410 reactions, n = 5 for 382–402, n = 4 for 367–404, and n = 3 for 367–402). The column labelled f.l. S6K1 shows the phosphorylation level of full-length S6K1ki (ki superscript indicates the kinase-inactive K100R mutant) under the same conditions, as reported in ref. 23. Truncation of the S6K1 peptide to 20 residues (S6K1382–402), which is the standardized length used in the peptide library of Fig. 1g, reduces phosphorylation to approximately 20% of S6K1367–404, probably in part because of end-effects destabilizing the helix as well as eliminating some minor contacts. b, Michaelis–Menten steady-state kinetic constants for mTORΔN–mLST8 (30 nM) phosphorylating wild-type and the indicated mutant S6K1367–404 peptides, quantified as in a. Graph shows means (dashes) and values (markers as indicated) from independent experiments (n = 3, except for the 1, 2, 10, 750, 900, and 1,200 μM points, which are n = 2). Also shown are the Km and kcat values, calculated by nonlinear regression fitting of the data, above the graph, and their simulated curves in the graph. Mutations that significantly reduce phosphorylation but do not make substantial direct FRB contacts include S394A, which eliminates a hydrogen bond that stabilizes the helix N terminus, and the helix-breaking V395G mutation, which further reduces phosphorylation compared with V395A (Fig. 1d); together, these point to the importance of the helical conformation. Additional mutations include Val391 and Pro393, in the segment between Thr389 and the start of the helix. Pro393 may be important for guiding the FRB-anchored substrate to the kinase active site, and that the P + 2 residue Val391 may be involved in contacts to the kinase C-lobe, where, by analogy with canonical kinases, the peptide segment of the phosphorylation site and its immediate vicinity are expected to bind. c, Superposition of the FRB–S6K1 interface onto the FRB–rapamycin–FKBP12 structure25 (FKBP12 is omitted for clarity), highlighting the similarities in the binding of the Leu396 side chain to the same pocket as rapamycin’s key C23 methyl group and the flanking portions of its triene arm. The FRB–S6K1 interface is coloured as in Fig. 1b; rapamycin is green and its associated FRB domain is cyan. In the crystals of the FRB–S6K1389–414 fusion protein, S6K1 residues 389–391 and 411–414 are disordered, while residues 405–410 are involved in crystal packing. d, Quantification of the reactions shown in Fig. 1h. e, X-ray data collection and refinement statistics for the FRB2018–2114-S6K1αII389–414 fusion protein structure.
Extended Data Figure 2 RAPTOR–4EBP1 TOS interface and density in the cryo-EM structure of mTORC1.
a, Human RAPTOR–4EBP1 TOS interface from the 3.0 Å refined human mTORC1 cryo-EM structure, coloured as in Fig. 2b. Only the RAPTOR side chains that make hydrogen bonds (red dotted lines) or electrostatic or van der Waals contacts to 4EBP1 are shown. b, Stereo view of the cryo-EM density of the human RAPTOR–4EBP1 TOS interface. Although the complex contained full-length 4EBP1, only an eight-residue segment is ordered in the cryo-EM reconstruction (also see Extended Data Fig. 7a). c, Cryo-EM data collection and refinement statistics.
Extended Data Figure 3 atRaptor–TOS motif crystal structures and human RAPTOR–TOS motif dissociation constants.
a, Stereo view of the mFo − dFc electron density of the atRaptor-4EBP199–118 co-crystals, calculated with phases from atRaptor before any 4EBP1 was built into the model. The structure is coloured as in Fig. 2b, and the map, calculated at 3.0 Å and contoured at 2.2σ, is shown as blue mesh. Of 20 residues in the 4EBP1 peptide in the crystals, only the 8-residue segment shown is ordered. b, Stereo view of the mFo − dFc electron density, calculated as in a, of the atRaptor-S6K11-14 co-crystals. The 3.1 Å map is contoured at 2.2σ. As with the 4EBP1 co-crystals, only 8 of the 14 S6K1 residues are ordered. c, Stereo view of the mFo − dFc electron density, calculated as in a, of the atRaptor–PRAS40124–139 co-crystals. The 3.35 Å map is contoured at 1.9σ. Only 8 of the 16 PRAS40 residues are ordered. In addition, side chain of Glu133 has poor density and is only tentatively built. d, Molecular surface representation of the atRaptor–TOS-binding groove, coloured according to the electrostatic potential (-6kT to +6kT) calculated without the 4EBP1 peptide (shown as sticks) using APBS54 and illustrated with PyMOL. The TOS-binding groove has an overall basic electrostatic potential due to five arginine and one lysine residues, explaining the tendency of acidic residues to be present at the non-conserved and flanking positions of the TOS motif. e, Binding of the indicated human 4EBP1 TOS mutant peptides (mutation in red) to human RAPTOR measured by fluorescence polarization anisotropy. Graph shows means as dashes and values from three independent experiments with the indicated markers and colours. Dissociation constants from the nonlinear regression fitting of the data are also shown, and simulated binding curves are overlaid on the data. f, Binding of the TOS motif peptides of human 4EBP1 (blue), S6K1 (red), and PRAS40 (green) to human RAPTOR measured by fluorescence polarization anisotropy. Graph shows means as dashes and values from three independent experiments with the indicated markers and colours. Also shown are the dissociation constants from the nonlinear regression fitting of the data and the simulated binding curves.
Extended Data Figure 4 PRAS40 is a competitive inhibitor of the FRB substrate-recruitment site and additionally binds to mLST8.
a, Inhibition of mTORΔN–mLST8 (30 nM) phosphorylating the TOS-less 4EBP142 (10 μM) by indicated PRAS40 fragments (red rectangles mark TOS, β-strand, and amphipathic α-helix). Incorporation of 32P is plotted as a fraction of the zero PRAS40 reaction of each series, with means as dashes and values from independent experiments with the indicated markers and colours (n = 3 for PRAS40206–256; n = 2 for PRAS40114–256, and PRAS40173–256). IC50 values from the nonlinear regression fitting of the data and their simulated curves are also shown. b, Co-crystal structure of the PRAS40173–256–mTORΔN–mLST8 complex. The mTOR FAT domain is coloured in cyan, FRB in salmon, kinase in pink, mLST8 in green, and PRAS40 in red. c, Anomalous diffraction Fourier map (red mesh) of crystals containing the SeMet-substituted L225M mutant of PRAS40206–256 bound to mTORΔN–mLST8 showing two SeMet peaks that confirm the direction of the PRAS40 helix. The 5.4 Å map is contoured at 3.5σ and superimposed on the structure of the wild-type complex, which has a slightly different unit cell from the SeMet crystals. The anomalous diffraction map of wild-type selenomethionine-substituted PRAS40 co-crystals is shown in the main text (Fig. 3b). d, The mFo − dFc electron density of PRAS40114–207, which contains only one of the three phenylalanine residues present in the PRAS40173–256 polypeptide, bound to mTORΔN–mLST8, confirming the sequence assignment of the PRAS40 β-strand. The map was calculated with phases from before PRAS40 was built into the model. The structure and map, calculated at 3.0 Å and contoured at 2σ, are coloured as in Fig. 3d. e, Phosphorylation of full-length 4EBP1 by apo-mTORC1 (left) does not obey Michaelis–Menten kinetics, and in the absence of a single substrate Km value we cannot calculate the Ki of full-length PRAS40 for the reaction of Fig. 3e. Left: the 32P incorporation data, plotted as velocity over enzyme concentration (means ± s.d., n = 11 except for the 10, 50, and 200 μM reactions, which are n = 6), for the curve of apo-mTORC1-phosphorylating full-length 4EBP1. The curve of reaction velocity/[enzyme] versus substrate concentration is parabolic, with product levels comparatively higher at low substrate concentrations (up to around 10 μM) and lower at higher substrate concentrations than a Michaelis–Menten-type response. The two substrate concentration ranges display very different Km and kcat values. The entire substrate range can thus be modelled (black curve) as the sum of two reactions, one having a tight Km of approximately 2 μM but very slow kcat of 0.003 s−1 (blue dashed curve), and another reaction with a weak Km of 545 μM but a faster kcat of 0.065 s−1 (green dashed curve). The tight Km reaction is dependent on the TOS motif, as its mutation (right; graph shows means as dashes and values from three independent experiments) results in a reaction that obeys Michaelis–Menten kinetics, with Km and kcat values of 462 μM and 0.053 s−1, respectively, which are very similar to the values of the weak Km faster kcat curve of wild-type 4EBP1. We presume that the TOS-independent weak Km reaction reflects, in part, substrate interactions with the FRB. It is possible that the non-Michaelis–Menten behaviour is due to the presence of multiple, probably non-equivalent phosphorylation sites on 4EBP1. We also cannot reliably measure the Km value of full-length S6K1, as this substrate aggregates and then precipitates at concentrations higher than approximately 50 μM, before reaching saturation (not shown). f, X-ray data collection and refinement statistics for the structures of mTORΔN–mLST8 bound to PRAS40 fragments.
Extended Data Figure 5 Cryo-EM reconstruction of mTORC1 and RHEB–mTORC1 complexes.
a, Flow chart of single particle cryo-EM data processing. Details are described in Methods. b, Left: gold-standard FSC plots between two independently refined half-maps for the consensus mTORC1 C2 reconstruction (red curve), the masked monomer with signal subtraction (blue curve), the RHEB–mTORC1 C2 reconstruction (purple curve), and the masked RHEB–mTORC1 monomer with signal subtraction containing additional signal-subtracted particles from a second, down-scaled dataset (green curve). The FSC cutoff of 0.143 and associated resolution for each plot are marked. Right: gold-standard FSC plots for the three focused refinements of the consensus mTORC1 C2 reconstruction with mask 1 (purple curve), mask 2 (red curve), and mask 3 (green curve) described in Methods. Also shown are the corresponding curves for RHEB–mTORC1 masks 1, 2, 3 in cyan, orange, and pink, respectively. The FSC cutoff of 0.143 and associated resolution ranges of the three masked refinements for consensus mTORC1 and RHEB–mTORC1 are marked on each plot. c, The four largest RHEB-less 3D classes of the flowchart (a) superimposed on the C-lobe of the kinase domain (marked by arrow) of one mTOR protomer. The closest approach of the two mLST8 subunits ranges from 123 Å (‘closed’ conformation in light blue) to 145 Å (‘open’ conformation in pink). The RHEB-containing class and two minor classes of suboptimal density are omitted for clarity. The four classes shown seem to represent samples along a continuum of conformations between the open and closed states, as more intermediate states get populated in a 3D classification with a larger number of classes (not shown). The 3D classes shown are from a calculation with a partial dataset approximately 50% the size of the final dataset. d, Comparison of the RHEB-containing class (green, with the RHEB density in red) with the RHEB-less classes (coloured light grey to dark grey). The maps are superimposed as in c. The RHEB-containing class has two RHEB molecules with very similar density, even though the 3D classification was done in C1. In the figure, one of the two RHEBs is occluded in the left and right panels, and is in lighter background (labelled as RHEB-2) in the middle panel. None of the RHEB-less classes has any significant density at either of the two RHEB-binding sites. The RHEB-containing class has an inter-mLST8 distance intermediate between the open and closed conformations of the RHEB-less classes, but the relative positions of N-heat and its associated RAPTOR are distinct from the apparent continuum of conformational states of the RHEB-less classes. Curved arrows indicate the transitions from the RHEB-less classes (‘minus’ sign) to the RHEB-containing (‘plus’ sign) class. e, Cryo-EM density of the RHEB-containing particles from the 3D classification showing cartoon representations of the refined RHEB–GTPγS structure (yellow, its switch I and II segments in red), and the RHEB-interacting portions of the mTOR N-heat (green), M-heat (pink), and FAT (pink) segments. f, FSC plots of the final model versus the composite cryo-EM map from REFMAC5 (black), and of a model validation protocol34 refining against one of two half-maps after an initial random displacement of atoms is applied to the model to remove model bias (FSCwork in red), and cross-validating the same model against the other half-map (FSCfree green).
Extended Data Figure 6 Secondary structure and conservation of mTOR.
Human mTOR sequence showing conservation from yeast to man (blue column graph above sequence) and secondary structure elements in the refined model. Helices are indicated as rectangles, β-strands as arrows, segments lacking regular secondary structure as solid lines, and disordered regions as dashed lines. N-heat secondary structure is coloured green, M-heat in orange, FAT in cyan (including helices fα1–fα6, which are continuous with the FAT structure, even though they are outside the FAT boundary defined by sequence conservation in PIKKs), the kinase domain N-lobe in yellow (FRB helices are named kfα1–kfα4 for consistency with the mTORΔN–mLST8 paper), and C-lobe in pink. The three hatched N-heat helices have not been assigned a sequence, and their boundaries are indicated tentatively. The dashed lines indicate disordered regions.
Extended Data Figure 7 Apo-mTORC1 cryo-EM density, interfaces, and conformational flexibility.
a, The 4EBP1 amphipathic helix density on the FRB does not have interpretable cryo-EM density. The map shown is of a 3D class (~30% of particles) with the highest relative level of density; the FRB side chains that contact the S6K1 substrate in Fig. 1c are shown as red sticks to map the site, the rest of mTOR is coloured as in Fig. 4a, and AMPPNP is in space-filling representation. Unlike the 4EBP1 TOS, which has a level of density comparable to that of its binding site (Extended Data Fig. 2b), the putative density of the 4EBP1 amphipathic helix is much weaker than that of the FRB, suggestive of partial occupancy. We presume this is due, in part, to our cryo-EM samples containing 260 mM NaCl, which substantially reduces 4EBP1 phosphorylation and thus probably further weakens substrate-FRB association (right). b, Stereo view of the 3.0 Å cryo-EM density of the consensus apo-mTORC1 reconstruction, showing mTOR N-heat (residues approximately 650–850 shown in stick representation coloured green, red, and blue for C, O, and N atoms, respectively). c, Stereo view of the 3.0 Å cryo-EM density of the consensus apo-mTORC1 reconstruction showing mTOR M-heat (residues approximately 960–1105 shown in stick representation coloured sand, red, and blue for C, O, and N atoms, respectively). d, The end of the N-heat solenoid (residues 848–898; green) is anchored on the middle of the FAT domain (residues 1565–1627; light cyan). Close-up view showing side chains (glycine Cα atoms as spheres) and backbone groups (blue spheres for amide and sticks for carbonyl groups) involved in intra-molecular van der Waals or hydrogen bonds (yellow dotted lines). For clarity, only N-heat residues 836–903 and FAT residues 1537–1664 are shown. e, The mTOR binding elements of RAPTOR (purple) are encompassed within its conserved RNC (raptor N-terminal conserved)7, with the caspase domain contacting M-heat of one mTOR protomer (sand), the caspase insertion contacting both M-heat and the N-heat (green) of the other mTOR protomer, and the first three armadillo repeats of its solenoid contacting N-heat. Side chains are shown as in d. Hydrogen bond contacts are shown as red dotted lines. N-heat and M-heat structural elements above the plane of the figure are omitted for clarity. f, The conformational flexibility of apo-mTORC1 is associated with bending at a major hinge region of three heat repeats (indicated by a box) in the N-heat solenoid, in between its FAT and RAPTOR–M-heat interacting segments. Figure shows Cα trace of the tripartite interface between N-heat of protomer 2, M-heat of protomer 1, and RAPTOR of the four apo-mTORC1 3D classes of Extended Data Fig. 5c (coloured as in Fig. 4a) and RHEB–mTORC1 (all red). The four apo-mTORC1 classes were refined to 4.5 Å or better, and together with RHEB–mTORC1 were superimposed on RAPTOR (residues 52–422). Figure also highlights the flexibility on the N-terminal half of the N-heat solenoid in apo-mTORC1 (see Supplementary Discussion).
Extended Data Figure 8 RHEB-induced conformational change in mTORC1.
a, Stereo view of the cryo-EM density from the 3.4 Å RHEB–mTORC1 reconstruction, showing the RHEB–mTOR interface in the same orientation and colouring as in Fig. 4c. The RHEB-interacting segments of mTOR are nα3–nα7 of N-heat, mα2–mα4 of M-heat, and fα2–fα3 of FAT. Most of the contacts made by RHEB are from its switch I and switch II regions, with a small number of additional contacts to N-heat and M-heat contributed by the nearby segments of residues 5–7 and 106–111. b, Stereo view of cryo-EM density from RHEB–mTORC1 (sand) and the 3.0 Å apo-mTORC1 (green), with the two structures and maps superimposed on the C-lobes as in Fig. 5d. c, AMPPNP (orange) cryo-EM density of apo-mTORC1. d, Steady-state kinetic analysis of mTORC1 phosphorylation of intact 4EBP1 in the presence of 250 μM RHEB–GTPγS. Reactions quantified by 32P incorporation and plotted as velocity over enzyme concentration (means as dashes and values from two independent experiments as filled circles). The Km and kcat values, calculated by nonlinear regression fitting of the data, and simulated curves are also shown. Note that in contrast to the reaction in the absence of RHEB shown in Extended Data Fig. 4e, the curve of reaction velocity versus 4EBP1 concentration obeys Michaelis–Menten kinetics. e, RHEB–GTPγS activation of 4EBP1 phosphorylation by mTORC1 under single-turnover conditions. A master mix of excess mTORC1 (500 nM) over 4EBP1 substrate (100 nM) was incubated with 250 μM RHEB–GTPγS or 250 μM RHEB–GDP in the standard kinase buffer on ice for 5 min. Reactions were started by the addition of a mixture of cold ATP (50 μM final) and [γ-32P]ATP (8 μCi per reaction time point). The reactions were done on ice to slow them down. At the indicated time points, an aliquot of the reaction was drawn, stopped, and analysed as described in Methods. The experiment was repeated three times with very similar results. f, ATP steady-state kinetic parameters of ATP hydrolysis by mTORC1 in the presence of 250 μM RHEB–GDP (left, blue plot) or RHEB–GTPγS (right, red plot). Reactions were quantified by 32P incorporation as in d. Graph shows means as dashes and values from three independent experiments with the indicated markers and colours. The steady-state kinetic constants of the RHEB–GDP-containing reaction are approximate owing to the weak signal of these reactions. g, As expected, RHEB–GTPγS did not activate the truncated mTORΔN–mLST8 complex phosphorylating 4EBP1 or S6K1367–404 (10 μM both; mTORΔN at 20 and 30 nM, respectively) (left; experiments were repeated twice with very similar results). mTORΔN–mLST8 has an intermediate kcat of 0.66 s−1 (Extended Data Fig. 1b) compared with the 0.09 and 2.9 s−1kcat values of apo-mTORC1 and RHEB–mTORC1, respectively (Fig. 5g). mTORΔN–mLST8 has a distinct FAT conformation, probably because of the absence of N-heat. Right: superposition of the FAT plus kinase domain portions of inactive apo-mTORC1 on the crystal structure of mTORΔN–mLST8 done by aligning their C-lobes. Apo-mTORC1 is in green and mTORΔN is coloured blue for FAT, yellow for N-lobe, and pink for C-lobe. The rotation axes (red lines) are numbered according to the hinges of Fig. 5c. Compared with the inactive-to-active transition, the comparison of the mTORΔN FAT conformation with that of the inactive state shows bigger changes around the major hinge with a rotation in the opposite direction and a different rotation axis far from the hinge axis (labelled ‘1’). The rotations around the two minor hinges are comparably modest although distinct, with the rotation axes nearly orthogonal to those of the inactive-to-active transition. h, Autoradiogram showing activation of mTORC1 phosphorylating 4EBP1 (10 μM) by RHEB–GTPγS, repeated three times. Gel quantification is shown in Fig. 5b. i, Steady-state kinetic analysis of mTORC1 phosphorylating S6K1367–404 in the presence of 250 μM RHEB–GDP (top row) or RHEB–GTPγS (bottom row). Incorporation of 32P data are plotted as velocity over enzyme concentration in Fig. 5g (n = 3).
Extended Data Figure 9 Hyperactivating mTOR mutations cluster in three regions of the mTOR structure.
a, Hyperactivating mutations that cluster at the major hinge region of the FAT domain. The mutations, which occur at residues with structure-stabilizing roles, probably disrupt the structural integrity of the FAT clamp and thus its ability to block the movement of the N-lobe into the active position. Cancer-genome mutations shown experimentally to be hyperactivating5 are mapped onto the apo-mTORC1 (left) and RHEB–mTORC1 (right) structures. The two structures are aligned on the C-lobes of their kinase domains to highlight the different packing arrangements at the hinge. The side chains of mutated residues are indicated with the letter ‘M’ and coloured dark cyan, the side chains that they interact with are in light cyan, while the rest of the structural elements are coloured as in Fig. 4a. Red dotted lines indicate groups within hydrogen bond distance. b, The largest cluster of hyperactivating mutations is centred on an N-lobe helical extension (kα3, kα3b) that is anchored in a pocket between the C-lobe and FAT domains. Here, the structural mutations either at the N-lobe kα3–kα3b helices or at their binding site on the C-lobe and the adjacent FAT domain would weaken the structural coupling of the N-lobe with the C-lobe, possibly allowing the N-lobe to assume conformations closer to its active state. Mutations mapped onto the apo-mTORC1 (left) and RHEB–mTORC1 (right) structures. c, Hyperactivating mutations that cluster where the FAT transitions into and packs with the N-lobe. Mutations here probably destabilize the structural elements and their packing which prevents the N-lobe from moving into its active position, mimicking the RHEB-induced conformational change and the associated looser FAT–N-lobe interface. Mutations are mapped onto apo-mTORC1 (FAT in cyan; N-lobe in yellow, C-lobe in pink) and RHEB–mTORC1 (all in grey). The two structures are superimposed on their N-lobe domains to facilitate comparison. d, Steady-state kinetic analysis of S6K1367–404 phosphorylation by 30 nM mTORC1 containing wild-type or the indicated hyperactive mTOR mutants. Incorporation of 32P was quantified and velocity over enzyme concentration values were plotted as means (dashes) and values from two independent experiments with the indicated markers and colours. Dissociation constants from the nonlinear regression fitting of the data are also shown, and simulated binding curves are overlaid on the data.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion sections about steady state kinetic constants, about other mTORC1 substrates in Figure 1g, about Raptor structure, about mTORC1 structure, and about apo-mTORC1 conformational flexibility. The file also contains Supplementary Figure 1 showing the source data (autoradiograms and immunoblots of gels) for the Main text and Extended Data figures. (PDF 17171 kb)
A morph from the inactive to the active mTOR conformation.
The roundtrip morph from the inactive (at video start) to the active (video middle) monomeric mTORC1 structures superimposed on the KD C lobes (mLST8 not shown). Orientation and coloring similar to Figure 4a. (MOV 1714 kb)
Close-up view of the catalytic cleft in the morph from the inactive to active mTOR conformation.
Close-up of Supplementary Video 1 focusing on the relative orientation of the kinase N and C lobes. View is similar to Figure 5d looking into the kinase catalytic cleft. (MOV 1566 kb)
Rights and permissions
About this article
Cite this article
Yang, H., Jiang, X., Li, B. et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 (2017). https://doi.org/10.1038/nature25023
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25023
This article is cited by
-
TFEB drives mTORC1 hyperactivation and kidney disease in Tuberous Sclerosis Complex
Nature Communications (2024)
-
mTORC1 in energy expenditure: consequences for obesity
Nature Reviews Endocrinology (2024)
-
TORC1 is an essential regulator of nutrient-controlled proliferation and differentiation in Leishmania
EMBO Reports (2024)
-
The rapid proximity labeling system PhastID identifies ATP6AP1 as an unconventional GEF for Rheb
Cell Research (2024)
-
Computational Analysis of Non-synonymous SNPs in ATM Kinase: Structural Insights, Functional Implications, and Inhibitor Discovery
Molecular Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.