Abstract
RNA has important and diverse roles in biology, but molecular tools to manipulate and measure it are limited. For example, RNA interference1,2,3 can efficiently knockdown RNAs, but it is prone to off-target effects4, and visualizing RNAs typically relies on the introduction of exogenous tags5. Here we demonstrate that the class 2 type VI6,7 RNA-guided RNA-targeting CRISPR–Cas effector Cas13a8 (previously known as C2c2) can be engineered for mammalian cell RNA knockdown and binding. After initial screening of 15 orthologues, we identified Cas13a from Leptotrichia wadei (LwaCas13a) as the most effective in an interference assay in Escherichia coli. LwaCas13a can be heterologously expressed in mammalian and plant cells for targeted knockdown of either reporter or endogenous transcripts with comparable levels of knockdown as RNA interference and improved specificity. Catalytically inactive LwaCas13a maintains targeted RNA binding activity, which we leveraged for programmable tracking of transcripts in live cells. Our results establish CRISPR–Cas13a as a flexible platform for studying RNA in mammalian cells and therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998)
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001)
Root, D. E., Hacohen, N., Hahn, W. C., Lander, E. S. & Sabatini, D. M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 (2006)
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003)
Tyagi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 6, 331–338 (2009)
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017)
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell 60, 385–397 (2015)
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016)
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017)
Dahlman, J. E. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 33, 1159–1161 (2015)
Hutchinson, J. N. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007)
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR–C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016)
Zetsche, B. et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Rath, S. et al. Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl Acad. Sci. USA 112, 15916–15921 (2015)
Gross, G. G. et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985 (2013)
Unsworth, H., Raguz, S., Edwards, H. J., Higgins, C. F. & Yagüe, E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 24, 3370–3380 (2010)
Nelles, D. A. et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165, 488–496 (2016)
Tourrière, H. et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003).
Tafer, H. et al. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 26, 578–583 (2008)
Mann, D. G. et al. Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236 (2012)
Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011)
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protocols 12, 828–863 (2017)
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651 (2006)
Bernhart, S. H., Hofacker, I. L. & Stadler, P. F. Local RNA base pairing probabilities in large sequences. Bioinformatics 22, 614–615 (2006)
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012)
Acknowledgements
We thank M. Alimova, D. Feldman, F. Chen, J. G. Doench, J. M. Engreitz, N. Habib, D. Tenen, A. Allen, R. Macrae, and R. Belliveau for discussions and support. O.A.A. is supported by a Paul and Daisy Soros Fellowship and a National Defense Science and Engineering Fellowship. J.S.G. is supported by a D.O.E. Computational Science Graduate Fellowship. A.R. is supported by the Howard Hughes Medical Institute. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. F.Z. is supported by the National Institutes of Health through the National Institute of Mental Health (5DP1-MH100706 and 1R01-MH110049), the Howard Hughes Medical Institute, the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; and James and Patricia Poitras, Robert Metcalfe, and David Cheng.
Author information
Authors and Affiliations
Contributions
O.O.A., J.S.G., and F.Z. conceived and designed the study. O.O.A. and J.S.G. participated in the design and execution of all experiments. P.E. performed biochemical characterization studies on the LwaCas13a protein. J.J. and V.V. prepared the vectors for arrayed knockdown screening experiments. J.J. also performed RNA immunoprecipitation experiments. D.B.T.C. assisted with cloning of constructs. M.J.K. performed the RNA integrity analysis. O.O.A. and J.S.G. analysed data. S.H. performed select microscopy experiments. J.J.B. performed the plant protoplast knockdown experiments. O.O.A., J.S.G., E.S.L., and F.Z. wrote the paper with input from D.F.V., A.Y.T., and A.R. and help from all authors.
Corresponding author
Ethics declarations
Competing interests
Patent applications have been filed relating to work in this manuscript. F.Z. is an adviser for Editas Medicine and Horizon Discovery.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
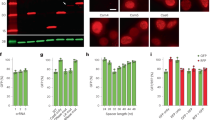
Extended Data Figure 1 Evaluation of LwaCas13a PFS preferences and comparisons with LshCas13a.
a, Sequence comparison tree of the 15 Cas13a orthologues evaluated in this study. b, Ratios of in vivo activity from Fig. 1b. c, Distributions of PFS enrichment for LshCas13a and LwaCas13a in targeting and non-targeting samples. The 25th and 75th percentiles are shown as grey dotted lines and the median is shown as a red dotted line. The minimum and maximum are marked by the ends of the distribution. Each distribution represents 976 PFS sequences (n = 976). d, Number of LshCas13a and LwaCas13a PFS sequences above depletion threshold for varying depletion thresholds. Values are mean ± s.e.m. with n = 2. e, Distributions of PFS enrichment for LshCas13a and LwaCas13a in targeting samples, normalized to non-targeting samples. The 25th and 75th percentiles are marked by the ends of the box and the median is shown as a red line within the box. Whiskers denote 1.5 times the interquartile range; blue ‘+’ denote outliers that are beyond 1.5 times the interquartile range. Each distribution represents 976 PFS sequences (n = 976). f, Sequence logos and counts for remaining PFS sequences after LshCas13a cleavage at varying enrichment cutoff thresholds. g, Sequence logos and counts for remaining PFS sequences after LwaCas13a cleavage at varying enrichment cutoff thresholds.
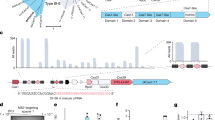
Extended Data Figure 2 Biochemical characterization of LwaCas13a RNA cleavage activity.
a, Gel electrophoresis comparison of LwaCas13a and LshCas13a RNase activity on ssRNA 1. b, Gel electrophoresis of ssRNA1 after incubation with LwaCas13a with or without crRNA 1 for varying amounts of times. c, Gel electrophoresis of ssRNA 1 after incubation with varying amounts of LwaCas13a–crRNA complex. d, Sequence and structure of ssRNA 4 and ssRNA 5. crRNA spacer sequence is highlighted in blue. e, Gel electrophoresis of ssRNA 4 and ssRNA 5 after incubation with LwaCas13a and crRNA 1. f, Sequence and structure of ssRNA 4 with sites of poly-x modifications highlighted in red. crRNA spacer sequence is highlighted in blue. g, Gel electrophoresis of ssRNA 4 with each of four possible poly-x modifications incubated with LwaCas13a and crRNA 1. h, Gel electrophoresis of pre-crRNA from the L. wadei CRISPR–Cas locus showing LwaCas13a processing activity. i, Cleavage efficiency of ssRNA 1 for crRNA spacer truncations after incubation with LwaCas13a. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 Engineering and optimization of LwaCas13a for mammalian knockdown.
a, Knockdown of Gluc transcript by LwaCas13a and Gluc guide 1 spacers of varying length. b, Knockdown of Gluc transcript with Gluc guide 1 and varying amounts of transfected LwaCas13a plasmid. c, Knockdown of Gluc transcript by LwaCas13a and varying amounts of transfected Gluc guide 1 and 2 plasmid (n = 2 or 3). d, Knockdown of Gluc transcript using guides expressed from either U6 or tRNAVal promoters (n = 2 or 3). e, Knockdown of KRAS transcript using guides expressed from either U6 or tRNAVal promoters (n = 2 or 3). f, Knockdown of KRAS and CXCR4 transcripts by LwaCas13a using guides transfected in A375 cells with position-matched shRNA comparisons (n = 2 or 3). g, Knockdown of Gluc transcript and endogenous transcripts PPIB, KRAS, and CXCR4 with active and catalytically inactive LwaCas13a. h, Validation of the top three guides from the arrayed knockdown Gluc and Cluc screens with shRNA comparisons (n = 2 or 3). i, Arrayed knockdown screen of 93 guides evenly tiled across the XIST transcript. All values are mean ± s.e.m. with n = 3, unless otherwise noted (n represents the number of transfection replicates). ***P < 0.001; **P < 0.01; *P < 0.05. n.s, not significant. A two-tailed Student’s t-test was used for comparisons.
Extended Data Figure 4 LwaCas13a targeting efficiency is influenced by accessibility along the transcript.
a, Top row: top knockdown guides are plotted by position along target transcript. Top knockdown guides are defined as the top 20% of guides for Gluc and the top 30% of guides for Cluc, KRAS, and PPIB. Bottom row: histograms for the pairwise distance between adjacent top guides for each transcript (blue) compared with a random null distribution (red). A shift of the blue curve (actual measured distances) to the left of the red curve (null distribution of distances) indicates that guides are closer together than expected by chance. b, Gluc, Cluc, PPIB, and KRAS knockdown partly correlates with target accessibility as measured by predicted folding of the transcript. The correlation was computed using a Pearson’s correlation coefficient and two-tailed Student’s t-test. c, Kernel density estimation plots depicting the correlation between target accessibility (probability of a region being base-paired) and target expression after knockdown by LwaCas13a. d, Top row: correlations between target expression and target accessibility (probability of a region being base-paired) measured at different window sizes (W) and for different k-mer lengths. Bottom row: P values for the correlations between target expression and target accessibility (probability of a region being base-paired) measured at different window sizes (W) and for different k-mer lengths. The colour scale is designed such that P values >0.05 are shades of red and P values <0.05 are shades of blue.
Extended Data Figure 5 Detailed evaluation of LwaCas13a sensitivity to mismatches in the guide–target duplex at varying spacer lengths.
a, Knockdown of KRAS evaluated with guides containing single mismatches at varying positions across the spacer sequence (n = 2 or 3). b, Knockdown of PPIB evaluated with guides containing single mismatches at varying positions across the spacer sequence (n = 2 or 3). c, Knockdown of Gluc evaluated with guides containing non-consecutive double mismatches at varying positions across the spacer sequence. The wild-type sequence is shown at the top with mismatch identities shown below. d, Collateral cleavage activity on ssRNA 1 and 2 for varying spacer lengths. e, Specificity ratios of guide tested in d. Specificity ratios are calculated as the ratio of the on-target RNA (ssRNA 1) collateral cleavage to the off-target RNA (ssRNA 2) collateral cleavage. f, Collateral cleavage activity on ssRNA 1 and 2 for 28-nt spacer crRNA with synthetic mismatches tiled along the spacer. g, Specificity ratios, as defined in e, of crRNA tested in f. h, Collateral cleavage activity on ssRNA 1 and 2 for 23-nt spacer crRNA with synthetic mismatches tiled along the spacer. i, Specificity ratios, as defined in e, of crRNA tested in h. j, Collateral cleavage activity on ssRNA 1 and 2 for 20-nt spacer crRNA with synthetic mismatches tiled along the spacer. k, Specificity ratios, as defined in e, of crRNA tested in j. For a–c, all values are mean ± s.e.m. with n = 3, unless otherwise noted (n represents the number of transfection replicates). For d–k, all values are mean ± s.e.m. with n = 4 (n represents the number of technical replicates).
Extended Data Figure 6 LwaCas13a is more specific than shRNA knockdown for endogenous targets.
a, Left: expression levels in log2(transcripts per million (TPM) + 1) values of all genes detected in RNA-seq libraries of non-targeting shRNA-transfected control (x axis) compared with KRAS-targeting shRNA (y axis). Shown is the mean of three biological replicates. The KRAS transcript data point is coloured in red. Right: expression levels in log2(transcripts per million (TPM) + 1) values of all genes detected in RNA-seq libraries of non-targeting LwaCas13a-guide-transfected control (x axis) compared with KRAS-targeting LwaCas13a-guide (y axis). Shown is the mean of three biological replicates. The KRAS transcript data point is coloured in red. b, Left: expression levels in log2(transcripts per million (TPM) + 1) values of all genes detected in RNA-seq libraries of non-targeting shRNA-transfected control (x axis) compared with PPIB-targeting shRNA (y axis). Shown is the mean of three biological replicates. The PPIB transcript data point is coloured in red. Right: expression levels in log2(transcripts per million (TPM) + 1) values of all genes detected in RNA-seq libraries of non-targeting LwaCas13a-guide-transfected control (x axis) compared with PPIB-targeting LwaCas13a-guide (y axis). Shown is the mean of three biological replicates. The PPIB transcript data point is coloured in red. c, Comparisons of individual replicates of non-targeting shRNA conditions (top row) and Gluc-targeting shRNA conditions (bottom row). d, Comparisons of individual replicates of non-targeting guide conditions (top row) and Gluc-targeting guide conditions (bottom row). e, Pairwise comparisons of individual replicates of non-targeting shRNA conditions against the Gluc-targeting shRNA conditions. f, Pairwise comparisons of individual replicates of non-targeting guide conditions against the Gluc-targeting guide conditions.
Extended Data Figure 7 Detailed analysis of LwaCas13a and RNAi knockdown variability (standard deviation) across all samples.
a, Heatmap of correlations (Kendall’s tau) for log2(transcripts per million (TPM + 1)) values of all genes detected in RNA-seq libraries between targeting and non-targeting replicates for shRNA or guide targeting either luciferase reporters or endogenous genes. b, Heatmap of correlations (Kendall’s tau) for log2(transcripts per million (TPM + 1)) values of all genes detected in RNA-seq libraries between all replicates and perturbations. c, Distributions of standard deviations for log2(transcripts per million (TPM + 1)) values of all genes detected in RNA-seq libraries among targeting and non-targeting replicates for each gene targeted by either shRNA or guide.
Extended Data Figure 8 LwaCas13a knockdown is specific to the targeted transcript with no activity on a measured off-target transcript.
a, Heatmap of absolute Gluc signal for first 96 spacers tiling Gluc. b, Heatmap of absolute Cluc signal for first 96 spacers tiling Gluc. c, Relationship between absolute Gluc signal and normalized luciferase for Gluc tiling guides. d, Relationship between absolute Cluc signal and normalized luciferase for Gluc tiling guides. e, Relationship between PPIB 2−Ct levels and PPIB knockdown for PPIB tiling guides. f, Relationship between GAPDH 2−Ct levels and PPIB knockdown for PPIB tiling guides. g, Relationship between KRAS 2−Ct levels and KRAS knockdown for KRAS guides. h, Relationship between GAPDH 2−Ct levels and KRAS knockdown for KRAS guides. i, Bioanalyzer traces of total RNA isolated from cells transfected with Gluc-targeting guides 1 and 2 or non-targeting guide from the experiment with active LwaCas13a in Extended Data Fig. 3g. The RNA integrity number (RIN) is shown and 18S rRNA and 28S rRNA peaks are labelled above. A Student’s t-test showed no significant difference for the RIN between either of the targeting conditions and the non-targeting condition. The curves are shown as a mean of three replicates and the shaded areas in light red around the curves show the s.e.m. j, The Bioanalyzer trace for the RNA ladder with peak sizes labelled above.
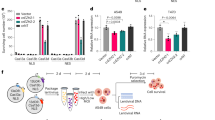
Extended Data Figure 9 dLwaCas13a–NF can be used for ACTB imaging.
a, Comparison between localization of dLwaCas13–GFP and dLwaCas13a–GFP–KRAB (dLwaCas13a–NF) constructs for imaging ACTB. Scale bars, 10 μm. b, Additional fields of view of dLwaCas13a–NF delivered with a non-targeting guide. Scale bars, 10 μm. c, Additional fields of view of dLwaCas13a–NF delivered with ACTB guide 3. Scale bars, 10 μm. d, Additional fields of view of dLwaCas13a–NF delivered with ACTB guide 4. Scale bars, 10 μm.
Extended Data Figure 10 dLwaCas13a–NF can image stress granule formation in living cells.
a, Representative images from RNA FISH of the ACTB transcript in dLwaCas13a–NF-expressing cells with corresponding ACTB-targeting and non-targeting guides. Cell outline is shown with a dashed line. Scale bars, 10 μm. b, Overall signal overlap between ACTB RNA FISH signal and dLwaCas13a–NF quantified by the Mander’s overlap coefficient (left) and Pearson’s correlation (right). Correlations and signal overlap were calculated pixel-by-pixel on a per cell basis; n = 10–25 cells per condition. ****P < 0.0001; ***P < 0.001; **P < 0.01. A two-tailed Student’s t-test was used for comparisons. c, Representative images from live-cell analysis of stress granule formation in response to 400 μM sodium arsenite treatment. Scale bars, 20 μm. d, Quantification of stress granule formation in response to sodium arsenite treatment. Quantification is based on overlapping dLwaCas13a–NF and G3BP1 puncta; n = 54–72 cells per condition. All values are mean ± s.e.m. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. n.s., not significant. A two-tailed Student’s t-test was used for comparisons.
Supplementary information
Supplementary Information
This file contains a discussion, notes 1-7 and references. (PDF 443 kb)
Supplementary Figure 1
This file contains extended data figures 2a, b, c, e, g, h, and i. (PDF 2397 kb)
Supplementary Table 1
This file contains the guides used for in vivo experiments in this study. (PDF 294 kb)
Supplementary Table 2
This file contains the plasmids used in this study. (PDF 108 kb)
Supplementary Table 3
This file contains the shRNA used in this study. (PDF 130 kb)
Supplementary Table 4
This file contains the ssRNA targets used in this study. (PDF 110 kb)
Supplementary Table 5
This file contains the guides used for in vivo experiments in this study. (PDF 111 kb)
Supplementary Table 6
This file contains the ssRNA targets and crRNAs used for the SHERLOCK experiments. (PDF 119 kb)
Supplementary Table 7
This file contains the guide sequences used in arrayed RNA knockdown screens. (XLSX 73 kb)
Supplementary Table 8
This file contains the Commercial TaqMan probes used in this study. (PDF 69 kb)
Supplementary Table 9
This file contains the Custom TaqMan probes used in this study. (PDF 62 kb)
Supplementary Table 10
This file contains the Cas13a orthologs used in this study. (PDF 120 kb)
Rights and permissions
About this article
Cite this article
Abudayyeh, O., Gootenberg, J., Essletzbichler, P. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017). https://doi.org/10.1038/nature24049
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24049
This article is cited by
-
Cas13b-mediated RNA targeted therapy alleviates genetic dilated cardiomyopathy in mice
Cell & Bioscience (2024)
-
Engineering a transposon-associated TnpB-ωRNA system for efficient gene editing and phenotypic correction of a tyrosinaemia mouse model
Nature Communications (2024)
-
Real-time single-molecule imaging of transcriptional regulatory networks in living cells
Nature Reviews Genetics (2024)
-
Transcript-specific induction of stop codon readthrough using a CRISPR-dCas13 system
EMBO Reports (2024)
-
Programmable RNA base editing with photoactivatable CRISPR-Cas13
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.