Abstract
In mammals, the canonical somatic DNA methylation landscape is established upon specification of the embryo proper and subsequently disrupted within many cancer types1,2,3,4. However, the underlying mechanisms that direct this genome-scale transformation remain elusive, with no clear model for its systematic acquisition or potential developmental utility5,6. Here, we analysed global remethylation from the mouse preimplantation embryo into the early epiblast and extraembryonic ectoderm. We show that these two states acquire highly divergent genomic distributions with substantial disruption of bimodal, CpG density-dependent methylation in the placental progenitor7,8. The extraembryonic epigenome includes specific de novo methylation at hundreds of embryonically protected CpG island promoters, particularly those that are associated with key developmental regulators and are orthologously methylated across most human cancer types9. Our data suggest that the evolutionary innovation of extraembryonic tissues may have required co-option of DNA methylation-based suppression as an alternative to regulation by Polycomb-group proteins, which coordinate embryonic germ-layer formation in response to extraembryonic cues10. Moreover, we establish that this decision is made deterministically, downstream of promiscuously used—and frequently oncogenic—signalling pathways, via a novel combination of epigenetic cofactors. Methylation of developmental gene promoters during tumorigenesis may therefore reflect the misappropriation of an innate trajectory and the spontaneous reacquisition of a latent, developmentally encoded epigenetic landscape.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013)
Ohm, J. E. et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 (2007)
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007)
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007)
Feinberg, A. P., Ohlsson, R. & Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7, 21–33 (2006)
Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017)
Schroeder, D. I. et al. The human placenta methylome. Proc. Natl Acad. Sci. USA 110, 6037–6042 (2013)
Branco, M. R. et al. Maternal DNA methylation regulates early trophoblast development. Dev. Cell 36, 152–163 (2016)
Deaton, A. M. & Bird, A. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2011)
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009)
Hon, G. C. et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206 (2013)
Ziller, M. J. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013)
Landan, G. et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat. Genet. 44, 1207–1214 (2012)
Landau, D. A. et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014)
Arman, E., Haffner-Krausz, R., Chen, Y., Heath, J. K. & Lonai, P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl Acad. Sci. USA 95, 5082–5087 (1998)
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013)
Boulard, M., Edwards, J. R. & Bestor, T. H. Abnormal X chromosome inactivation and sex-specific gene dysregulation after ablation of FBXL10. Epigenet. Chromatin 9, 22 (2016)
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008)
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014)
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015)
MacLeod, A. R., Rouleau, J. & Szyf, M. Regulation of DNA methylation by the Ras signaling pathway. J. Biol. Chem. 270, 11327–11337 (1995)
Lu, C. W. et al. Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat. Genet. 40, 921–926 (2008)
Serra, R. W., Fang, M., Park, S. M., Hutchinson, L . & Green, M. R. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife 3, e02313 (2014)
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010)
Walter, M. J. et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25, 1153–1158 (2011)
Rhee, I. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556 (2002)
Lin, H. et al. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol. Cell. Biol. 26, 2976–2983 (2006)
Novakovic, B. & Saffery, R. Placental pseudo-malignancy from a DNA methylation perspective: unanswered questions and future directions. Front. Genet. 4, 285 (2013)
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011)
Smith, Z. D. et al. DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615 (2014)
Chenoweth, J. G. & Tesar, P. J. Isolation and maintenance of mouse epiblast stem cells. Methods Mol. Biol. 636, 25–44 (2010)
Smith, Z. D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012)
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013)
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014)
Yoshida, N. & Perry, A. C. Piezo-actuated mouse intracytoplasmic sperm injection (ICSI). Nat. Protocols 2, 296–304 (2007)
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008)
Ying, Q. L., Nichols, J., Chambers, I. & Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 (2003)
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013)
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016)
Macaulay, I. C . et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015)
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols 9, 171–181 (2014)
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protocols 6, 468–481 (2011)
Wu, H. et al. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 43, e141 (2015)
Ben-Porath, I. et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008)
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009)
Schep, A. N. et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015)
Ciruna, B. G. & Rossant, J. Expression of the T-box gene Eomesodermin during early mouse development. Mech. Dev. 81, 199–203 (1999)
Ralston, A. & Rossant, J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614–629 (2008)
Savory, J. G. et al. Cdx2 regulation of posterior development through non-Hox targets. Development 136, 4099–4110 (2009)
Donnison, M. et al. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132, 2299–2308 (2005)
Goldin, S. N. & Papaioannou, V. E. Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis 36, 40–47 (2003)
Kang, M., Piliszek, A., Artus, J. & Hadjantonakis, A. K. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140, 267–279 (2013)
Nichols, J., Silva, J., Roode, M. & Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222 (2009)
Auclair, G., Guibert, S., Bender, A. & Weber, M. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 15, 545 (2014)
Smallwood, S. A. et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 (2011)
Ooi, S. K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007)
He, J. et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 15, 373–384 (2013)
Wu, X., Johansen, J. V. & Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 49, 1134–1146 (2013)
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 (2014)
Boulard, M., Edwards, J. R. & Bestor, T. H. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 47, 479–485 (2015)
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009)
Steine, E. J. et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J. Clin. Invest. 121, 1748–1752 (2011)
Schulze, I. et al. Increased DNA methylation of Dnmt3b targets impairs leukemogenesis. Blood 127, 1575–1586 (2016)
Yang, L. et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell 29, 922–934 (2016); erratum 30, 363–365, (2016)
Mayle, A. et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125, 629–638 (2015)
Haney, S. L. et al. Promoter hypomethylation and expression is conserved in mouse chronic lymphocytic leukemia induced by decreased or inactivated Dnmt3a. Cell Rep. 15, 1190–1201 (2016)
Acknowledgements
We thank members of the Meissner and Michor laboratories for discussions and advice, in particular R. Karnik for help with data processing and alignment, as well as B. E. Bernstein and R. P. Koche for their expertise. F.M. and J.S. gratefully acknowledge support from the Dana-Farber Cancer Institute Physical Sciences-Oncology Center (NIH U54CA193461). The work was funded by the New York Stem Cell Foundation, the Broad-ISF Partnership for Cell Circuit Research, the Starr Foundation, NIH grants (1P50HG006193, P01GM099117 and R01DA036898) and the Max Planck Society. A.M. is a New York Stem Cell Foundation Robertson Investigator.
Author information
Authors and Affiliations
Contributions
Z.D.S., J.S., F.M. and A.M. designed and conceived the study and prepared the manuscript. Z.D.S. performed all experiments and assisted in data analysis as performed by J.S. J.D. made the ATAC–seq libraries, D.C. made RNA-seq libraries, and H.G. made the dual RRBS and RNA-seq libraries with supervision from A.G. and alignment by K.C. F.M. and A.M. jointly supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
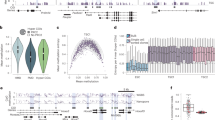
Extended Data Figure 1 Tracking divergence in DNA methylation landscapes during mouse implantation.
a–f, Sequencing metrics and coverage information for WGBS, RNA-seq, and ATAC–seq data including hierarchical clustering and Pearson correlation for CpGs, genes, and gene promoters, respectively. WGBS data also includes Euclidean distance, which can be beneficial for examining sample similarity in globally hypomethylated samples, as well as similarity scores for 100 bp tiles, which locally merge the intrinsically higher variance of intermediately methylated CpGs to reduce noise. For RNA-seq and ATAC–seq data, biological replicates cluster together, as do 8-cell and postimplantation WGBS data, whereas tissues of the E3.5 blastocyst cluster together but not as discrete inner cell mass (ICM) and trophectoderm (TE) compartments. In general, there is minimal variation between the methylation status of the ICM and trophectoderm, with only slight deviations around the minimal global value that is reached during this developmental period. g, Isolation of the epiblast and ExE from the E6.5 post-implantation embryo. The conceptus is first removed from maternal decidual tissue and portioned into epiblast and ExE fractions, taking care to remove the apical ectoplacental cone (EPC). Then, outer visceral endoderm (VE) and trophoblast cells are enzymatically digested and mechanically removed using a thin glass capillary.
Extended Data Figure 2 Unique features of the extraembryonic methylation landscape.
a, CpG methylation boxplots for all covered CpGs as well as those that are significantly hyper- or hypomethylated within the ExE compared to epiblast (ExE hyper or ExE hypo, respectively). ExE hypo CpGs largely reflect differential remethylation compared to the epiblast across the genome. Alternatively, ExE hyper CpGs are mostly unmethylated in the ICM and trophectoderm and remain so in the epiblast, indicating an ExE-specific mechanism. Edges refer to the 25th and 75th percentiles, and whiskers the 2.5th and 97.5th percentiles, respectively. b, Differential methylation distribution for ExE hyper or hypo CpGs compared to epiblast. Hypomethylation appears to be a global feature of the ExE and deviates from a default hypermethylated state in the epiblast. Alternatively, increased DNA methylation appears to be directed focally and de novo at regions that are unmethylated within the epiblast and subsequent embryonic and adult somatic tissues. c, Alternate CpG density distributions for ExE hypo and hyper CpGs indicate differential enrichment within distinct genomic features. Whereas ExE hypo CpGs resemble the global average, ExE hyper CpGs occur within regions of higher CpG densities. d, The fraction of dynamically methylated CpGs that fall within annotated exons as a function of distance to their assigned TSS. 44% of exonal ExE hyper CpGs fall within 2 kb of their associated TSS. e, The fraction of dynamically methylated CpGs that fall within annotated CpG islands (CGIs) based upon their proximity to the nearest TSS. ExE hyper CpGs are generally TSS proximal and skew downstream of the TSS, with 43% falling within ±2 kb. f, DNA methylation distribution for different genomic features including those associated with genic (TSS, exon, intron and CGI) and repetitive (LINE, SINE and LTR) sequences. For reference, black bar and arrows highlight the global median and the 25th and 75th percentiles, respectively. Globally, all features exhibit the expected passage through minimal DNA methylation values within the ICM and trophectoderm of the E3.5 blastocyst before remethylation at implantation. Compared to its global distribution, the ExE exhibits higher levels of de novo methylation within exons and introns, and lower than global levels within regions of LINE and LTR retrotransposon origin. The epiblast exhibits nearly complete hyper or hypomethylation depending on the genomic feature, and is bimodal at TSSs, which frequently contain CGIs. n values refer to the number of annotated features of a given type. g, Violin plots of 100 bp methylation data for early embryonic, placental, and fetal tissues demonstrate general epigenetic retention of either the somatic epiblast or extraembryonic architecture throughout subsequent development. The white dot highlights the global median, and blue and red reflect the median of ExE hypomethylated 100-bp tiles and ExE hyper CGIs, respectively. Notably, the placenta largely preserves the hypomethylated global landscape and targeted CGI methylation as they are established by E6.5 within the ExE. We show 100-bp tiles and CGIs for ExE-specific hypomethylation and hypermethylation, respectively, to restrict CpGs to a notable feature where they change as a group. WGBS data of somatic tissues and midgestation placenta taken from ref. 11.
Extended Data Figure 3 Transcriptional differences between epiblast and ExE are directed in part through DNA methylation.
a, Select gene set enrichment analysis of ExE hypermethylated TSSs, including Gene Ontology, canonical pathways, and genetic and chemical perturbations, shows high enrichment for transcription factors and signalling pathways involved in patterning the early embryo. Moreover, these promoter CGI-containing genes are canonical targets of PRC2, which coordinates selective expression of key developmental regulators during gastrulation. b, DNA methylation and open chromatin dynamics for the tumour suppressors p16Ink4a, p19Arf (both encoded by Cdkn2a) and p15Ink4b (encoded by Cdkn2b). Although these loci are either basally or non-transcribed during early development, three regions are dynamically methylated in the ExE (highlighted in grey), including a >10-kb region that encompasses the entirety of the p16Ink4a locus and is either wholly unmethylated in the epiblast or extensively methylated in the ExE. CGIs are highlighted in green, and the positions of included TSSs are highlighted in red. c, Scatterplot of log2 expression dynamics versus differential CGI methylation between the epiblast and the ExE. Although most dynamically methylated CGI promoter-containing genes have functions in later embryonic development and are not yet highly expressed, de novo methylation in the ExE is generally associated with transcriptional repression. ExE hyper CGIs are highlighted in pink. Promoter CGIs are assigned to the most proximal gene within a boundary of ±2 kb. d, Boxplots relating promoter methylation and expression in the restriction of extraembryonic and embryonic compartments. Promoters are defined as ±1 kb of an annotated TSS and scored as dynamically methylated in the ExE if the difference with the epiblast is ≥0.1. Expression changes between dynamically methylated and background promoter sets are provided over increasing thresholds according to their expression in the epiblast. Although many CGI promoter-containing genes are not dynamically expressed in either the epiblast or the ExE and are associated with downstream developmental functions, transcriptional repression is a consistent feature of promoter methylation, even at this low threshold. e, Median open chromatin signal as measured by ATAC–seq for ExE hyper CGI-associated TSSs in the transition from pre- to postimplantation. ExE hyper CGI-associated genes are heavily enriched for roles in patterning the embryo proper and are primarily not expressed until the onset of gastrulation. In the transition from blastocyst to epiblast, these promoters gain open chromatin signal, suggesting transcriptional priming or activation, which is not observed within the ExE, where they are de novo methylated. Shaded area reflects the 25th and 75th percentile per fixed 100-bp bin. f, Expression and differential promoter methylation of key epigenetic and master transcriptional regulators over early embryonic and extraembryonic development. Most epigenetic regulators exhibit minimal expression differences between the epiblast and the ExE, with the Dnmts being notable exceptions. Key isoforms of Dnmt3a and Dnmt3b are upregulated in the epiblast in conjunction with global remethylation, whereas the suppression of Dnmt3a in the ExE corresponds with de novo promoter methylation. Alternatively, the maintenance methyltransferase Dnmt1 and the non-catalytic cofactor Dnmt3l are induced within the blastocyst and maintained at higher levels in the ExE, with reciprocal methylation of the Dnmt3l promoter in the epiblast. The H3K36 demethylase Kdm2b displays differential expression of catalytically active and inactive isoforms within the epiblast and the ExE, respectively, with isoform switching seemingly imposed by de novo methylation around the somatically used CGI promoter. The ExE is characterized by persistent expression of the master regulators Cdx2, Eomes, and Elf5 (refs 48, 49, 50, 51), whereas the still pluripotent epiblast remains Pou5f1 (also known as Oct4) positive. Many additional regulators of subsequent developmental stages are basally expressed within the epiblast and their promoters de novo methylated in the ExE. The difference in promoter methylation refers to the annotated TSS that exhibits the greatest absolute difference between ExE and epiblast. TPM, transcripts per million. Additional high-resolution genome browser tracks are displayed for select transcriptional and epigenetic regulators in Extended Data Figs 4 and 7, respectively. g, Unsupervised hierarchical clustering of 11,780 genes over late preimplantation and early post-implantation development, partitioned into 20 distinct dynamics (‘clusters’). Cluster 10 includes genes that are specifically induced within the epiblast but not the ExE. Heat map intensity reflects the row-normalized z-score. h, Significant Gene Ontology enrichment for the 20 gene expression dynamics characterized in f, including those regulated by ExE-methylated CGI promoters, as calculated using the binomial test. Cluster 10 is enriched for both developmental functions and ExE promoter methylation.
Extended Data Figure 4 Unique bifurcation and epigenetic reinforcement of transcriptional regulators during postimplantation development.
a, Genome browser tracks for WGBS, ATAC–seq and RNA-seq data for transcriptional regulators associated with embryonic or extraembryonic development. CGIs are highlighted in green, and the positions of included TSSs are highlighted in red. Embryonic regulators include Pou5f1, Nanog, and Pdrm14, which are progressively expressed over preimplantation and for which Pou5f1 and Nanog remain expressed in the epiblast. For these genes, repression in the ExE is accompanied by differential methylation of their TSSs, which is apparent as a local hypermethylation ‘peak’ at the Pou5f1 locus within an ~5 kb region that is otherwise hypomethylated in the epiblast. At the Nanog locus, an upstream region remains hypomethylated in both tissues. Finally, de novo methylation of the Prdm14 promoter is representative of ExE-specific CGI promoter methylation that occurs at hundreds of genes with downstream developmental functions. Density refers to the projected number of methylated CpGs per 100 bp of primary sequence and highlights the extensive epigenetic signal present over these regions within the ExE specifically (Δdensity refers to the difference compared to the epiblast). b, Extraembryonic development is in part directed by the master regulator Elf5, which is not induced until implantation and is reciprocally methylated at its TSS in the epiblast. Intriguingly, many transcriptional regulators associated with pluripotency and germline development persist within the ExE, including Zfp42 and the paralogues Dppa2 and Dppa4. As with Elf5, the promoters for these genes are differentially methylated in the epiblast and frequently characterized by broad kilobase-scale hypomethylation surrounding their TSSs in the ExE. c, Scatterplots for log2 TPM as a function of promoter methylation reveal a higher sensitivity to low methylation levels in the ExE in comparison to the epiblast. Median, 25th, and 75th percentiles for expression within 0.1 methylation bins are included for reference. The fraction of unmethylated promoters is very similar between each tissue and exhibit comparable expression values. Promoters are calculated as ±1 kb of an annotated TSS. Vertical dotted line indicates the median methylation value of ExE hyper CGIs. d, Read-level methylation of ExE hyper CGIs in the ExE and epiblast. The methylation status for every sequencing read within a given CGI was ranked and binned into percentiles. Plotted are the median and the 25th and 75th percentiles for these ranks across ExE hyper CGIs for both the ExE and the epiblast. In general, about 80% of reads falling within these regions are methylated in the ExE, with a median methylation value of 0.25. This value is very close to the average, unphased measurement for the CGI entirely, indicating that de novo methylation occurs in a high fraction of cells within the ExE and to a similar extent.
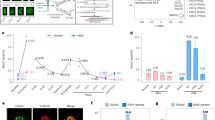
Extended Data Figure 5 Epigenetic restriction of FGF production and sensing to embryonic or extraembryonic compartments.
a, Genome browser tracks for WGBS, ATAC–seq and RNA-seq data for select growth factors, receptors, and potentiators that are dynamically regulated during early post-implantation development. Fgf loci such as the ICM-expressed Fgf4 and epiblast-expressed Fgf5 and Fgf8 are all regulated by CGI-containing promoters that are de novo methylated in the ExE. Alternatively, expression of FGF-sensing genes such as Fgfr2 and the potentiating protein Fgfbp1 becomes specific to the ExE and is characterized by broad kilobase-scale hypomethylated domains surrounding their respective TSSs in this tissue. Moreover, the asymmetric allocation of Fgfr2-expressing cells during the specification of the ICM indicates that this tissue is still sensitive to these growth factors before the epigenetic restriction that is imposed by DNA methylation during implantation52,53. CGIs are highlighted in green, and the positions of included TSSs are highlighted in red. Density refers to the projected number of methylated CpGs per 100 bp of primary sequence and highlights the extensive epigenetic signal present over these regions within ExE specifically (Δdensity refers to the difference compared to epiblast). b, Bright-field images of ICM outgrowths after two or four days under disparate growth factor or small molecule conditions. All ICMs were cultured on irradiated feeders in a basal N2/B27 media supplemented with leukaemia inhibitory factor (LIF). 2i refers to the canonical FGF-inhibited, WNT-active condition comprised of the MEK inhibitor PD0325901 and the GSK3β inhibitor CHIR99021, which functions as a WNT agonist37. PD refers to culture with PD0325901 alone and represents repressed FGF signalling in the absence of an additional WNT input54. FGF4/CHIR represents dual FGF and WNT activity by culture in recombinant FGF4 and CHIR99021 and includes notable interior and exterior tissue structures that emerged during culture and were independently isolated and profiled. Finally, ICMs were cultured in FGF4 alone. Outlines highlight the specific components of each outgrowth that were subsequently purified for analysis by dual RNA-seq and RRBS profiling (see Methods). Scale bar shown on the bottom right. c, Differential methylation of CGIs during in vitro culture differs from the ExE according to developmental trajectory. Shown are specific TSS-associated CGIs that are either methylated in the ExE and both conditions, ExE and FGF/CHIR, or ExE-only and the corresponding mean adjusted log2 fold change in gene expression. Shared targets include early developmental genes, such as Prdm14, that are repressed in each case, though often highly expressed in the FGF/CHIR interior. Notably, some of these genes, particularly those associated with the germline, can be de novo methylated later in embryonic development55. FGF differs from the ExE and FGF/CHIR conditions in the methylation of CGIs associated with either the epiblast or the neuroectoderm, including genes that are expressed in the FGF condition, such as Otx2, Igfbp2, and Sfrp2, though this set encompasses other neuroectodermal master regulators such as Pax6 that are not yet expressed. Finally, ExE and FGF/CHIR diverge in the promoter methylation of endodermal master regulators, such as Foxa2, Hnf1b, Gata4, and Sox17, which are highly expressed in the transition from FGF/CHIR inside to outside. Notably, the bifurcation in CGI methylation corresponds to the expression of Fgfr2 and repression of Fgf4, as is observed in vivo: Fgf4 is highly expressed within the interior and repressed in the exterior (32.0 to 3.5 TPM) while Fgfr2 is induced (2.3 to 13.5 TPM). PD and FGF/CHIR conditions are also uniquely positive for Dnmt3b and 3l expression, but ExE hyper CGI methylation is not observed with PD0325901 present (TPM = 30.2 and 60.9 for Dnmt3b and Dnmt3l in FGF/CHIR outside, and 61.0 and 41.3 for PD), indicating either the requirement for an additional cofactor or post-translational modification to redirect these enzymes to this feature set.
Extended Data Figure 6 Generation of dual expression and methylation libraries from outgrowth and embryonic knockout data.
a, b, Sequencing metrics and coverage information for dual RNA-seq and RRBS libraries generated for the evaluation of ICM outgrowths and CRISPR–Cas9 disrupted E6.5 embryos, including similarity metrics between replicates (Euclidean distance and Pearson correlation for RRBS and Pearson correlation for RNA-seq). Mean and median methylation of 100 bp tiles is also included for the RRBS samples. c, CRISPR–Cas9 disrupted embryos were generated by zygotic injection of three single guide RNA (sgRNA) sequences specific to early exons that are shared across different isoforms. The genomic coordinates and protospacer sequences are provided (see Methods).
Extended Data Figure 7 Dynamic behaviour of key epigenetic regulators during early implantation.
Genome browser tracks for WGBS, ATAC–seq and RNA-seq data (log2 TPM shown for selected isoforms). CGIs are highlighted in green, and the positions of included TSSs are highlighted in red. a, Dnmt1 is not appreciably expressed in early cleavage, in part owing to a transient maternal imprint over the somatically used TSS (Dnmt1s)33,56, but shows moderate induction within the ICM. Then, at implantation, it is induced within both the epiblast and the ExE. Dnmt1 is expressed at higher levels within the ExE and displays persistent focal hypomethylation around the maternal-specific TSS (Dnmt1o) that is not observed in the epiblast, which resolves an area of preimplantation-specific hypomethylation to the hypermethylated genomic average. b, The short Dnmt3a2 isoform is induced to high levels in epiblast and is also expressed within embryonic stem (ES) cells. Alternatively, the CGI-containing promoter of Dnmt3a2 is methylated in the ExE and its transcription is suppressed. c, Like Dnmt1, the Dnmt3b promoter contains a CGI that is maternally imprinted during preimplantation33,56. Induction is apparent within the blastocyst, but becomes asymmetrically abundant within the epiblast following implantation. d, DNMT3L is a non-catalytic cofactor that enhances the de novo activity of DNMT3A and B, with specific functions in the early embryo and germline57. During implantation, Dnmt3l is initially expressed in both the ICM and the trophectoderm, but it remains expressed in the ExE and is silenced by de novo promoter methylation in the epiblast. e, The H3K36 demethylase KDM2B has specific roles in establishing the boundary between promoters and actively transcribed gene bodies, as well as in PRC2 recruitment and the establishment of facultative heterochromatin58,59,60,61. A catalytically inactive isoform, Kdm2b2, initiates from an alternate TSS downstream of exons encoding the demethylating Jumonji domain of the catalytically active Kdm2b1 (ref. 17). Kdm2b2 is the most prevalent isoform during preimplantation development and remains expressed in the ExE. Alternatively, Kdm2b1 is only induced during implantation within the Epiblast, whereas its CGI-containing promoter gains methylation in the ExE. Like Dnmt1s and Dnmt3b, the CGI promoter of Kdm2b1 is a maternally methylated imprint that resolves to hypomethylation during implantation33,56. f, Extraembryonic genome remethylation is highly dependent on DNMT3B and DNMT1. Pairwise comparisons of 100-bp tiles as measured by RRBS for wild-type epiblast and ExE (y axis) versus matched CRISPR–Cas9-disrupted tissues (x axis). Extraembryonic methylation levels diminish genome-wide when Dnmt1, Dnmt3b and Dnmt3l are disrupted. The epiblast is only sensitive to Dnmt1 and Dnmt3b disruption, both to a lesser extent than the ExE, presumably because of compensation from DNMT3A. Notably, the decrease in global methylation levels when Dnmt1 is deleted is greater for ExE than epiblast, indicating a higher dependence on maintenance and less efficient de novo methyltransferase activity in this tissue. The identity line is included in grey and the best fit by LOESS regression in red. The number of 100 bp tiles used in each comparison and the r2 values are included in the upper left of each plot. g, Composite plots of ExE hyper CGI-containing promoters in CRISPR–Cas9 targeted epiblast and ExE, respectively. In general, only limited effects are observed in the epiblast other than a slight increase in the peripheral methylation within the Eed-null sample. Alternatively, both TSS proximal and peripheral methylation is decreased in Dnmt1-, Dnmt3b-, and Dnmt3l-null ExE. The Eed-null ExE is unique in its specificity for diminished methylation at the TSS, particularly downstream within the first kilobase. In both the epiblast and the ExE, the wild-type median is included in black for comparison. Line represents the median and the shaded area the 25th and 75th percentiles, respectively. For RRBS data, composite plots are of the median for 200-bp windows, taken at intervals of 50 bp. h, Statistical test for the derepression of ExE hyper CGI associated genes demonstrates a comparable requirement for Eed in both the epiblast and the ExE. Gene expression of knockout samples were compared to matched wild-type samples using DESeq2 with raw counts as input. Enrichment for ExE hyper CGI associated genes were evaluated by Wilcoxon rank-sum test and represented as z-scores, which were converted to P values assuming a normal distribution. Bonferroni correction for multiple testing was applied to derive the FDR.
Extended Data Figure 8 General features of the cancer methylome and of CGI DMRs.
a, Median methylation of differentially regulated CGI-containing promoters in a primary colon tumour isolate and CLL compared to colon and B lymphocytes, respectively, as measured by WGBS. ExE hyper CGIs as identified in this study and shown in Fig. 1 are included for reference. The median methylation difference between extraembryonic or cancerous tissue compared to the epiblast or normal tissue is also included. The general features of both cancer methylomes are similar to those of the ExE, with a maximal increase in DNA methylation centred at the TSS that steadily diminishes within the periphery. Alternatively, hypomethylated CGIs in extraembryonic or tumorigenic contexts are maximally different a distance away from the TSS, within the boundary or ‘CpG island shore’, as previously reported for cancer62. Shaded area represents the 25th and 75th percentiles per 100-bp bin. b, Read-level methylation of hypermethylated CGIs in the ExE versus the epiblast, colon tumour versus colon, and CLL versus B lymphocyte, with those that share differential methylation status between the cancer and extraembryonic development included as a subset. The methylation status for every sequencing read within a given hypermethylated CGI was ranked and binned into percentiles. Plotted are the median and 25th and 75th percentiles for these ranks across CGIs called as hypermethylated in each pairwise comparison. The ExE/epiblast and CLL/B lymphocyte comparisons exhibit very similar distributions that indicate general discordance, meaning similar aggregate methylation across the feature as is observed in phase, which is most likely to be obtained by dispersive de novo methylation across the majority of alleles within the population. Colon tumour exhibits substantially higher read-level methylation, with a median of ~0.7. However, the per-read methylation level of the non-tumorous, matched colon tissue is also quite high, with >50% of reads exhibiting some methylation. This could indicate a transition in the epigenetic status of these loci within colon tissue that precedes tumorigenesis, as has been noted for several other tissues in Extended Data Fig. 9. The read-level methylation distribution is the same for cancer type-specific CGIs regardless of whether or not they are also ExE hyper CGIs. As such, the targeting to ExE hyper CGIs is a conserved feature of human cancer types, but the extent to which they are methylated can be specific to the system. c, Data taken from ENCODE samples that reflect embryonic and extraembryonic identities in human in comparison to the well-characterized human cancer cell line HCT116. The human ES cell line HUES64, a proxy for the pluripotent epiblast, displays notable enrichment for both repressive, PRC2-deposited H3K27me3 and activating H3K4me3 modifications at orthologous ExE hyper CGIs. Alternatively, human placenta exhibits diminished enrichment for both modifications at these regions, as does HCT116. Both systems display substantial methylation over ExE hyper CGIs as presented in Fig. 4, Extended Data Fig. 9 and Supplementary Table 7. As a control, ‘ExE hypo’ CGIs demonstrate uniformly high H3K4me3 levels. Enrichment density heat maps are provided for the full ExE hyper CGI set and are ranked across plots according to their enrichment for H3K27me3 in HUES64. Normalized enrichment represents the fold chromatin immunoprecipitation-enrichment against sample matched whole cell extract (WCE). d, Boxplots of mean methylation for 489 ExE-methylated, orthologous CGIs (ExE hyper CGIs) across the 14 tissue-matched TCGA cancer types that display disregulated DNA methylation landscapes and for CLL. Asterisk: CLL samples were measured by RRBS (n = 119) and represent a comparison between age-matched healthy B lymphocytes (n = 24). Edges refer to the 25th and 75th percentiles, whiskers the 2.5th and 97.5th percentiles, respectively. e, Boxplots for TCGA datasets and CLL for the absolute methylation values of all orthologously mapped CGIs, those methylated across cancer types, and those that are specifically methylated in mouse ExE. In all 15 cancer types that exhibit general global hypomethylation and CGI methylation as part of their departure from somatic cells, ExE hyper CGIs are specifically enriched, more so than for CGIs that are observed as hypermethylated in any given cancer type. f, Boxplots for the same data for cancer type-specific CGI DMRs and those that are also methylated in mouse ExE. Notably, the extent to which mouse ExE hyper CGIs are methylated reflects the cancer type, with some exhibiting higher absolute methylation values than others. However, in 14 out of 15 cases, the absolute methylation status of cancer type-specific CGI DMRs and those that are also methylated in the ExE are nearly identical, and often slightly greater. Absolute methylation values therefore appear to be determined by the specific cancer or cancer type, whereas targeting of extraembryonically methylated CGIs is a general feature.
Extended Data Figure 9 Broad conservation of extraembryonic methylation patterns across cancer types and cell lines.
a, Boxplots of orthologous ExE hyper CGIs across 107 ENCODE/Roadmap Epigenomics Project samples as presented in Fig. 4, with notable additional features of each sample highlighted below. Human extraembryonic tissues, including a trophoblastic cell line and primary placenta, also share conserved CGI methylation with mouse. Normal tissues that appear to exhibit higher mean methylation of ExE hyper CGIs include numerous endodermal lineages, such as colonic mucosa, stomach and liver (mean methylation values of 0.225, 0.185 and 0.179, respectively) as well as mature cell types of the adaptive immune system, such as CD8+ and CD4+ T lymphocytes and B lymphocytes (mean methylation values of 0.199, 0.173 and 0.173, respectively). By contrast, ectodermal and epithelial cells are comparatively less methylated than other somatic tissues, although cancer cell lines and primary tumours derived from these tissues remain sensitive to hypermethylation. b, Genome browser tracks for orthologous loci as originally presented for mouse development in Fig. 1 for three human fetal tissues that represent each germ-layer (brain, ectoderm; heart, mesoderm; stomach, endoderm), primary human B lymphocytes, and a CLL sample. CGIs around these loci are preserved in a hypomethylated state during embryonic development, where the bimodal architecture of the DNA methylation landscape is clearly maintained. In B lymphocytes, some low-level, encroaching methylation is already apparent over developmentally hypomethylated regions, as is also observed in the Roadmap sample in a. However, in the transition to CLL, extensive methylation is observed across these CGIs although methylation values drop in the surrounding areas. Red line and shaded area reflect the local mean and standard deviation as calculated by local regression (LOESS) to compensate for the greater number of CpGs within the human orthologues versus mouse, which can complicate visual estimates of local methylation at these scales. CGIs are highlighted in green.
Extended Data Figure 10 Genetic features of ExE CGI methylation in cancers.
a, Intersection analysis as presented in Fig. 4d for cancer-hypomethylated CGIs across the 14 TCGA cancer types and CLL that exhibit global loss of methylation in tandem with CGI hypermethylation. Generally, CGI hypomethylation is more specific, such that the intersection across cancers decays exponentially. Notably, even for hypomethylated CGIs, the intersection across cancer types remains higher for those that are also hypomethylated in mouse ExE, human placenta, or both (Conserved). b, Intersection analysis for cancer-dysregulated genes across TCGA cancer types. Of genes significantly dysregulated in at least n (0–14) TCGA cancer types, the fraction of genes that are functionally related to ExE hyper CGI-associated genes were predicted by GRAIL, using a global gene network built by text-mining (see Methods). An FDR of 5% was used as a cut-off. As the number of TCGA cancer types increases, the fraction of ExE hyper CGI-associated genes within the downregulated set generally increases, whereas those that are upregulated decrease substantially. c, Boxplots of the average methylation for the 489 orthologous ExE hyper CGIs across the 10,629 cancers available in TCGA with matched mutational and methylation data, segregated by mutational status of genes that function as part of the FGF signalling pathway. In aggregate, cancers with FGF pathway mutations have a median average ExE hyper CGI methylation level of 0.328 compared to 0.275 for those that do not (P < 10−16, rank-sum test). Edges refer to the 25th and 75th percentiles, whiskers the 2.5th and 97.5th percentiles, respectively. d, Among 539 genes that are present in the top 10 recurrently mutated pathways in cancer, 68 are functionally related to ExE hyper CGI-associated genes (FDR < 5%), as predicted by GRAIL. Genes in the FGF signalling pathway are highlighted in red. In general, FGF signalling pathway genes have high connectivity scores to ExE hyper CGI-associated genes (enrichment z-score = 3.88 for FGF pathway members within the P value distribution for all 539 genes). e, Statistical enrichment for FGF pathway genes for either amplification or deletion within the TCGA database is notably skewed towards amplification, indicating a generally oncogenic nature for this pathway in tumorigenesis. f, Methylation status of ExE hyper CGIs across colonic and haematopoietic mouse cancer models in which de novo methyltransferase activity has been perturbed. All samples are measured by RRBS. Datasets include: primary colon tissue in which Dnmt3b has been overexpressed (promoter methylation status reported, ref. 63); genetic models of acute myeloid leukaemia (AML) including those transformed by the MLL–AF9 fusion (ref. 64), cMyc and BCL2 overexpression (ref. 64), and FLT3 internal tandem duplication (FLT3-IDT, ref. 65); and acute and chronic lymphoblastic leukaemia models driven by Dnmt3a knockout alone (refs 66 and 67). Methylation of ExE hyper CGIs is observed in both colonic Dnmt3b overexpression and haematopoietic Dnmt3a knockout. Additional oncogenic drivers appear to induce de novo methylation of these regions in the presence or absence of DNMT3 expression, indicating numerous potentials routes to accomplishing the same molecular phenotype. Wild-type haematopoietic tissues are included for reference and taken from refs 66 and 67. Edges refer to the 25th and 75th percentiles, whiskers the 2.5th and 97.5th percentiles, respectively.
Supplementary information
Supplementary Table 1
De novo methylation of CGIs during extraembryonic development. Methylation status of CGIs in Epiblast and Extraembryonic Ectoderm (ExE), including designation of differential methylation status in ExE as described in the Methods (hyper, hypermethylated; hypo, hypomethylated; NC, no change; ND, insufficient measurements). Assignment to nearest gene and distance to the TSS are included. (XLSX 1711 kb)
Supplementary Table 2
Promoter methylation and associated transcriptional dynamics during implantation are influenced by CGI methylation status. Methylation values for gene promoters (classified as the region +/– 1 kb of an annotated TSS), Log2 normalized TPM (Transcripts per Million) across late preimplantation and early postimplantation samples. Promoter methylation is reported if at least 5 CpGs are covered ≥5x. The 'Symbol' column identifies all annotated genes for a given promoter and the reported expression value is either the TPM of the associated gene or the mean TPM if multiple genes begin at the same TSS. 'CpGs' indicates the number of CpGs that exist within the promoter boundary. (XLSX 4984 kb)
Supplementary Table 3
CGI methylation status for ICM outgrowths under defined conditions. CGI methylation status as measured by RRBS for ICM explanted under conditions of modulated FGF and WNT signaling. CGIs are assigned to their nearest TSS and those existing within +/– 2 kb were given the additional assignment of TSS-associated. DMR status indicates differential methylation between Epiblast and ExE from WGBS data, and PRC2 regulatory status is taken from Ref 67. We observe three discrete scenarios where CGIs are preferentially methylated: within the ExE, in the external portion of FGF+CHIR stimulated ICM outgrowths, and in FGF stimulated outgrowths. A CGI whose methylation status deviates by ≥0.1 from epiblast is scored as 'dynamic' and used to generate the heatmap in Fig. 2f. (XLSX 2147 kb)
Supplementary Table 4
Promoter methylation status and transcriptional dynamics for ICM outgrowths under defined conditions.Promoter methylation and associated gene expression data of ICM outgrowth conditions as measured by dual RRBS and RNA-seq. Promoter methylation is reported if at least 5 CpGs are covered ≥5x. The 'Symbol' column identifies all annotated genes for a given promoter and the reported expression value is either the TPM of the associated gene or the mean TPM if multiple genes begin at the same TSS. (XLSX 5274 kb)
Supplementary Table 5
CGIs methylation status for epigenetic regulator deficient E6.5 embryos. CGI methylation status as measured by RRBS for samples isolated from CRISPR/Cas9 injected embryos. CGIs are assigned to their nearest TSS and those existing within +/– 2 kb were given the additional assignment of TSS-associated. DMR status indicates differential methylation between Epiblast and ExE from WGBS data, and PRC2 regulatory status is taken from Ref 67. A CGI whose methylation status deviates by ≥0.1 from its wild type tissue is scored as 'dynamic' and is highlighted in Fig. 3d. (XLSX 2806 kb)
Supplementary Table 6
Promoter methylation status and transcriptional dynamics for epigenetic regulator deficient E6.5 embryos. Promoter methylation and associated gene expression data of CRISPR/Cas9 targeted embryos as measured by dual RRBS and RNA-seq. Promoter methylation is reported if at least 5 CpGs are covered ≥5x. The 'Symbol' column identifies all annotated genes for a given promoter and the reported expression value is either the TPM of the associated gene or the mean TPM if multiple genes begin at the same TSS. In general, ExE Hyper CGIs are preferentially induced in both the Epiblast and ExE fraction of Eed targeted E6.5 embryos and de novo methylation of these regions in ExE is specifically blocked. (XLSX 8983 kb)
Supplementary Table 7
Methylation status of ExE hypermethylated CGIs within human tissues, cancers, and cell lines. Mean and median methylation status of the 489 orthologously mapped CGIs that are called as ExE-hypermethylated in mouse across 107 ENCODE and Roadmap Initiative samples. Note, the lymphoblastoid cell line GM12878 is not characterized as cancer cell line within Encode but was generated using the Epstein-Barr Virus and scored as such in this study. Information includes designation as cancer versus normal as well as other assignments included in Extended Data Figure 9. (XLS 61 kb)
Rights and permissions
About this article
Cite this article
Smith, Z., Shi, J., Gu, H. et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549, 543–547 (2017). https://doi.org/10.1038/nature23891
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23891
This article is cited by
-
Important role of DNA methylation hints at significant potential in tuberculosis
Archives of Microbiology (2024)
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Self-patterning of human stem cells into post-implantation lineages
Nature (2023)
-
Spatiotemporal transcriptomic maps of whole mouse embryos at the onset of organogenesis
Nature Genetics (2023)
-
Mechanisms and function of de novo DNA methylation in placental development reveals an essential role for DNMT3B
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.