Abstract
For many enveloped viruses, binding to a receptor(s) on a host cell acts as the first step in a series of events culminating in fusion with the host cell membrane and transfer of genetic material for replication1,2. The envelope glycoprotein (Env) trimer on the surface of HIV is responsible for receptor binding and fusion. Although Env can tolerate a high degree of mutation in five variable regions (V1–V5), and also at N-linked glycosylation sites that contribute roughly half the mass of Env, the functional sites for recognition of receptor CD4 and co-receptor CXCR4/CCR5 are conserved and essential for viral fitness. Soluble SOSIP Env trimers are structural and antigenic mimics of the pre-fusion native, surface-presented Env3,4, and are targets of broadly neutralizing antibodies. Thus, they are attractive immunogens for vaccine development5,6,7,8. Here we present high-resolution cryo-electron microscopy structures of subtype B B41 SOSIP Env trimers in complex with CD4 and antibody 17b, or with antibody b12, at resolutions of 3.7 Å and 3.6 Å, respectively. We compare these to cryo-electron microscopy reconstructions of B41 SOSIP Env trimers with no ligand or in complex with either CD4 or the CD4-binding-site antibody PGV04 at 5.6 Å, 5.2 Å and 7.4 Å resolution, respectively. Consequently, we present the most complete description yet, to our knowledge, of the CD4–17b-induced intermediate and provide the molecular basis of the receptor-binding-induced conformational change required for HIV-1 entry into host cells. Both CD4 and b12 induce large, previously uncharacterized conformational rearrangements in the gp41 subunits, and the fusion peptide becomes buried in a newly formed pocket. These structures provide key details on the biological function of the type I viral fusion machine from HIV-1 as well as new templates for inhibitor design.
Similar content being viewed by others
Main
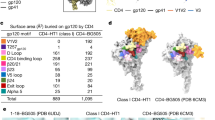
Numerous biophysical studies have provided a framework for CD4-induced Env conformational changes and dynamics through a combination of low-resolution cryo-electron tomography of membrane-embedded trimers9,10, X-ray crystallography of gp120 monomers11, and more recently hydrogen/deuterium-exchange mass spectrometry12 and Förster resonance energy transfer13 experiments (Supplementary Discussion). To study the molecular basis of conformational changes resulting from binding of receptors to HIV-1 Env, we obtained a cryo-electron microscopy (cryo-EM) map of fully glycosylated B41 SOSIP.664 (a solubilized and stabilized version of Env)14 in complex with two-domain soluble CD4 (sCD4) and the CD4-induced antibody 17b at 3.7 Å resolution (B41CD4–17b) (Fig. 1a, Extended Data Fig. 1, Extended Data Table 1). We also obtained a 5.6 Å resolution cryo-EM map of ligand-free subtype B B41 SOSIP.664 (B41LF) and showed it to be structurally comparable to subtype A BG505 (Extended Data Fig. 2c–f). Like BG505 SOSIP.664, B41 SOSIP.664 is a stable, native-like trimer that induced autologous neutralizing antibody responses in rabbit immunization studies15 but can adopt slightly more open conformations (partially open) when analysed by negative-stain electron microscopy, and has a lower melting temperature14,16 compared to BG505 (Extended Data Fig. 2a, b). Despite differences in their biophysical properties, the structure of B41LF (Extended Data Fig. 2f) adopts a closed, pre-fusion conformation nearly identical to the analogous BG505 SOSIP.664 structure (Protein Data Bank (PDB) accession number: 4ZMJ)17.
a, Cryo-EM map segmented by component. b, CD4 binding results in a number of conformational changes in both the gp120 and gp41 regions of Env. Comparison of the CD4–17b-bound (middle) to pre-fusion states (side panels).
Our structure of B41CD4–17b elucidates the molecular details of receptor-induced conformational changes, including rearrangements of the V1/V2 and V3 loops, and previously unobserved changes in gp41, including repositioning of the fusion peptide (Fig. 1b, Supplementary Video 1). In our B41CD4–17b cryo-EM map that contains the complete gp120, most of V1/V2 that extends parallel to CD4 is disordered in the region that was truncated in the core gp120 construct used for crystallography (residues C131 to N187) (Extended Data Fig. 3b). Despite the presence of 17b, the epitope of which overlaps the co-receptor binding site, much of the V3 loop beyond the base is also disordered.
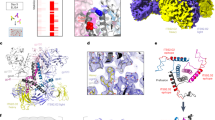
CD4–17b binding induces subtle changes in a network of conserved residues (>96% of sequences in the Los Alamos database (https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html)) in the gp120 core similar to previous observations in gp120 monomers11,18,19,20 (Fig. 2a, Extended Data Table 2, Supplementary Video 2). These changes propagate across gp120 to the C1 region (residues 63–72; α0), which is poorly ordered in pre-fusion Env trimer structures (Figs 1b, 2c). α0 adopts a stable α-helical conformation via transit into a pocket that was sterically occluded by the close juxtaposition of gp120 and HR1 in gp41 in the closed, pre-fusion trimer (Extended Data Fig. 3d, Supplementary Video 3). An additional rearrangement of two mannose residues in the D1 arm of the highly conserved N262 glycan into a new pocket underneath the base of V3 further stabilizes the helical conformation of α0 and establishes a direct link between the co-receptor binding site and gp41 (Fig. 2c, Extended Data Fig. 4c). Owing to the rotation of gp120, α0 now caps the HR1 helix (α7) of the gp41 subunit from a neighbouring protomer, which was previously capped by α1 in gp120 from within a protomer (Fig. 2b). This functional role of α0 is in contrast to a recently published low-resolution structure of a closed conformation Env trimer in complex with CD4, which suggested that CD4 makes specific contacts with C1 in the adjacent protomer21. In the open CD4-bound structure presented here, all of the highly conserved residues proposed to interact with CD4 during initial binding instead form intra-gp120 stabilizing interactions (Fig. 3d). Mutation of these residues lead to a loss of infectivity21; in light of the present work, this was probably due to destabilization of the conformation competent for co-receptor binding.
a, A network of hydrophobic and aromatic residues transmits the signal upon sCD4 binding that leads to the structural rearrangements in both gp120 and gp41. b, The top of the HR1 helix is capped by the α1 helix of the same protomer in the pre-fusion state (left). CD4-binding results in the formation of helix α0, which now caps HR1 of a neighbouring protomer (middle). A close-up of the α0–HR1 interaction reveals that it is downstream to the engineered I559P mutation and probably a feature in native Env (right). c, The D1 arm of glycan N262 rotates into a newly formed pocket on gp120 (left) positioned under the co-receptor binding site at the base of V3, providing a link between the co-receptor binding site and gp41. The α0 helix region is generally disordered in all published crystal structures of SOSIP trimers as evidenced by higher B-factors for this segment relative to the average gp120 value for each structure (right).
a, Space-filling model of B41CD4–17b illustrating a newly created pocket that is now occupied by the fusion peptide. A close-up view of the pocket (centre) and space-filling model (right). b, K574 of gp41, which forms a salt bridge with D107 of gp120 in the pre-fusion state, now interacts with F53 of gp120. c, The tryptophan clasp, previously described as a stabilizing feature of the pre-fusion state14, is retained in the CD4-bound state. d, Intra-protomer stabilizing interactions of helix α0 and β3–β4. e, Two aromatic residues of the fusion peptide rearrange upon CD4-binding and fit in two pockets. Sequence alignment of the fusion peptides of BG505 (pre-fusion) and B41 (CD4-bound) with the aromatic residues highlighted. All pre-fusion model coordinates are based on PDB: 5CEZ.
We produced additional cryo-EM reconstructions to test the effect of sCD4 alone on Env conformational changes, and compare with a different Env genotype, BG505 (Extended Data Fig. 1d). Within the limits of resolution, we observed only slight differences that were dependent on whether B41 or BG505 SOSIP was used, and whether 17b was present or not (Extended Data Figs 3c, 4d). The V3 loop remains unresolved in reconstructions of B41 or BG505 SOSIP in complex with sCD4, suggesting that sCD4 alone is responsible for the gp120 rearrangements and exposure of the co-receptor site, and suggesting that 17b does not induce further conformational changes (Extended Data Fig. 4f).
VRC01-class antibodies22 bind the CD4 binding site but do not induce global conformational changes in the Env trimer, whereas another CD4-binding-site neutralizing antibody, b12, has been shown to engage Env in a different manner9,14,23. To better understand the conformational changes induced by b12, we generated cryo-EM structures of B41 SOSIP.664 in complex with either CD4-binding-site antibody PGV04 (B41PGV04) or b12 (B41b12) to resolutions of 7.4 Å and 3.6 Å, respectively (Fig. 4a, Extended Data Figs 1, 5a, c). Although B41PGV04 was nearly identical to BG505 SOSIP.664 in complex with VRC01-like antibodies, B41b12 promoted a large-scale change in Env away from the closed pre-fusion conformation similar to B41CD4–17b (Extended Data Fig. 5b, d, e). Specifically, B41b12 exhibits a large rigid body movement of all three gp120 subunits with respect to the trimer axis and concomitant rearrangement of the gp41 helices (discussed below) (Fig. 4c, Extended Data Fig. 5d). Despite the gp120 rearrangements, the V3 loop remains sequestered beneath V1/V2 and the co-receptor binding site is inaccessible (Fig. 4b, Extended Data Fig. 6b). Although b12 makes similar interactions with Env as does CD4, it does not induce a conformational change in V1/V2 relative to gp120 (Fig. 4b, Extended Data Fig. 4e). Notably, the subtle rearrangement of mannose residues in the N262 glycan observed in B41CD4–17b is not seen in B41b12, and the α0 region is disordered. Thus, functional progression of the Env requires both a rigid body movement of gp120 and CD4-induced allosteric changes within gp120 to expose the co-receptor binding site. Unlike CD4, b12 is sterically occluded from binding the closed pre-fusion conformation of Env (Extended Data Figs 4b, 6c–f), suggesting that transient opening of the trimer is required for b12 epitope exposure. Upon binding, b12 prevents reversion back to the closed pre-fusion state (Extended Data Fig. 6d).
a, Cryo-EM map segmented by component. b, Comparison of gp120 variable loops before and after b12 binding; B41LF (left) and B41b12 (middle). Superposition of gp120 from pre-fusion and b12-bound states reveals only slight structural differences in the individual gp120 subunits (right). r.m.s.d., root-mean-square deviation. c, Structural rearrangements in gp41 between the pre-fusion and b12-bound states include repacking of the HR1 three-helix bundle as well as ordering of the fusion peptide and fusion peptide proximal region. Alignment onto HR2 of single gp41 chains from B41LF and B41b12 reveals a relative translocation of HR1 and rearrangement of the fusion peptide and fusion peptide proximal region (right).
Globally, both CD4–17b and b12 result in gp41 rearrangements that, in the case of the former, prime for further transitions to facilitate fusion with the host cell membrane. The HR1 three-helical bundle begins to open up at the base, causing the N-terminal half of the HR1 peptide (HR1N) to move away from the centre of the trimer (Extended Data Fig. 7g). This movement coincides with formation of a stable helix in the fusion peptide proximal region and HR1N that packs against HR2 (Extended Data Fig. 7h). Additionally, the gp120 C1 (V36–P43) and C5 (starting at I491) regions undergo a similar movement in the same direction to avoid clashing with α9b of HR2. Finally, HR2 tilts upward and, when modelled onto the surface of a virion, would be more perpendicular to and lifted off the membrane compared to the pre-fusion state.
One of the major consequences of the movement of gp120 relative to gp41 is the formation of a new pocket close to the trimer core that, in B41b12 and B41CD4–17b, houses the fusion peptide. The fusion peptide, which is fully resolved to the N-terminal residue A512 in B41b12 and L515 in B41CD4–17b, undergoes a large conformational rearrangement upon entry into this pocket (Fig. 3a, e, Extended Data Fig. 7c–e, Supplementary Video 4) and is stabilized by many newly formed interactions. For example, K574, which forms a salt bridge with D107 of gp120 in the closed pre-fusion state, undoes this interaction upon translocation of α1 induced by CD4–17b-binding, but forms a cation-π interaction with gp120 F53 (Fig. 3b, Extended Data Fig. 7a). All three residues (F53, D107 and K574) are >99% conserved and the interactions between them probably contribute to two stable states (Extended Data Table 2). Mutational analysis demonstrates that introduction of an F53A mutation not only decreases infectivity but also leads to greater gp120 shedding18,24. The gp120 interface with gp41 α7 in B41CD4–17b is further stabilized by highly conserved residues that do not make any productive interactions in the pre-fusion state (Fig. 3b). Finally, despite the large conformational change, conserved phenylalanine residues in the fusion peptide and fusion peptide proximal region provide further stabilizing interactions (Fig. 3e). In addition to formation of new contacts, the intermediate state exemplified by B41CD4–17b also retains some stabilizing features of the closed pre-fusion state (Fig. 3c, Extended Data Fig. 7f). Taken together, this extensive network of interactions between highly conserved residues suggests that the CD4–17b-bound state represents a stable, fusion intermediate wherein the fusion peptide is embedded inside the trimer distant from the host membrane and therefore requires further triggering by co-receptor engagement.
The Env trimer on the surface of HIV must maintain a balance between stability until a target cell is reached and the ability to trigger and undergo the marked conformational rearrangement that drives fusion. Our studies describe Env, even when stabilized by SOSIP mutations, as a dynamic molecule that fluctuates between closed and open pre-fusion states. In the open conformation, we now see that the fusion peptide is structured and sequestered near to the core of the trimer. We hypothesize that the fusion peptide may transition between solvent-exposed and sequestered conformations based on ‘breathing’ in the trimer. The structure of the fusion peptide in our b12- or CD4–17b-bound structures could therefore be used to inform structure-based design of stabilized or cyclic fusion peptide mimetic inhibitors of HIV25,26.
Finally, we show that CD4-binding-site neutralizing antibodies can prevent exposure of the co-receptor binding site in both the open (for example, b12) or closed conformation (for example, VRC01) by restricting movement of the V1/V2 loops (Extended Data Fig. 8). CD4, on the other hand, induces a series of allosteric changes that propagate across gp120 and expose the co-receptor binding site. The co-receptor binding site is accessible in the CD4-bound structure, but it is unclear how further conformational changes in the trimer are triggered by interaction with CCR5/CXCR4, although the glycan at N262 and formation of α0 probably have a role in signal transmission. Owing to the disposition of the co-receptor binding sites, three copies of CCR5 could bind to the trimer in this conformation27. Notably, CCR5 forms a crystallographic dimer in the X-ray structure27 that when docked on top of our trimer structure spans across two gp120 subunits and could potentially interact with two co-receptor binding sites simultaneously (Extended Data Fig. 8). Overall, our structural studies have unlocked the molecular details of the first step in the CD4-receptor-mediated fusion process of Env, although the role of the co-receptor requires further structural investigation.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Protein expression
BG505 SOSIP.664 and B41 SOSIP.664 trimers were expressed by transient transfection (co-transfected with furin) in HEK293F (Invitrogen; mycoplasma-free) and purified as previously described using 2G12-affinity and size-exclusion chromatography4,14.
The two N-terminal domains of human CD4 were cloned into vector pHCMV3 with an N-terminal Igκ secretion signal and a C-terminal hexahistidine tag for expression in HEK293F. Subsequently, 1 l of 293 FreeStyle cells was transiently transfected with 0.5 mg of DNA. After 7 days, the supernatant containing the secreted protein was applied to a 5 ml HisTrap FF column (GE Healthcare) at 1 ml min−1. The protein was eluted with a gradient from 0–100% of 1.5 M glycine in 0.02 M Tris-acetate pH 8.0, and 0.5 M NaCl. Peaks containing CD4 (as verified by SDS–PAGE) were pooled and further purified by size-exclusion chromatography on an S75 16/60 column (GE Healthcare) in 0.05 M Tris-acetate pH 8.0 and 0.3 M NaCl.
PGV04, 17b or b12 Fab was expressed in HEK293F and purified by 5 ml KappaSelect or LambdaSelect affinity (GE Healthcare), MonoS 5/50 GL (GE Healthcare), and Superdex 200 Increase 10/300 GL (GE Healthcare).
Sample preparation
BG505 SOSIP.664 were mixed with a 10× molar excess (sCD4:trimer) of sCD4 for 2 h at room temperature. For complexes containing 17b, a subsequent incubation was performed with a 10× molar excess (Fab:trimer) of 17b Fab for 1 h at room temperature. The complex was purified by size exclusion chromatography (SEC) using Superose 6 10/300 GL (GE Healthcare) in TBS (0.05 M Tris pH 7.4, 0.15 M NaCl). The fractions containing the complex were pooled and concentrated using a 100-kDa concentrator (Amicon Ultra, Millipore) to around 40 μl at 1 mg ml−1. 5 μl of the complex was incubated with 3 μl of a fresh DDM solution at 1.8 mM. A 3-μl aliquot of the complex was applied to a C-Flat grid (CF-2/2-4C, Electron Microscopy Sciences, Protochips, Inc.), which had been plasma-cleaned for 5 s using a mixture of Ar/O2 (Gatan Solarus 950 Plasma system), blotted off, and then immediately plunged into liquid ethane using a manual freeze plunger.
The same procedure was used to prepare the B41 complexes except that the incubation time of trimer and ligand(s) was increased to around 18 h before SEC. This long incubation time increased stoichiometry of binding for ligands with slow on-rates and did not have an effect on overall trimer stability judged by the lack of dimers or monomers in both SEC peaks and electron microscopy classifications.
Cryo-EM data collection
Frozen-hydrated samples were inserted into a Titan Krios electron microscope (FEI, Hillsboro, Oregon) operating at 300 kV. Data was collected through the Leginon software solution28,29,30. Each micrograph movie was collected at a magnification of 22,500, which resulted in a pixel size of 1.31 Å in the specimen plane. Micrograph movies were acquired using a K2 direct detector camera (Gatan) operated in counting mode with 200 ms exposure per frame. Dose rate was around 10 e− per pixel per s and defocus range was −1.0 to −4.0 μm. Micrograph movie frames were aligned and dose-weighted using MotionCor2 (ref. 31). CTF models were calculated using GCTF32.
Data processing for B41LF, B41b12, B41PGV04 and B41CD4–17b
Molecular projection image candidates were then identified in the aligned micrograph movies using DoG Picker33. Reference-free, 2D classification was performed using identified projection image candidates binned by a factor of two via iterative multivariate statistical analysis and multi-reference alignment34,35. Particles corresponding to class averages of the molecular complexes were selected for further processing in Relion version 2.036. Initially, full-size molecular projection images were refined against a simulated density map of an unliganded BG505 SOSIP Env trimer (simulated from the Env part of PDB: 5CEZ) low-pass filtered at 60 Å. Initial refinement was followed by 3D classification and stable symmetrical classes were selected for further symmetrized (C3) refinement. Resolutions were calculated using soft-edged masks that encompassed the entire structure (including all noisier regions) and are reported according to the FSC 0.143 gold-standard criterion. Final resolutions are ~3.6 Å (B41b12), ~3.7 Å (B41CD4–17b), ~5.2 Å (B41CD4), ~5.6 Å (B41LF) and ~7.4 Å (B41PGV04).
Data processing of BG505sCD4 and BG505sCD4–17b
Single particles were selected using DoG Picker33 from the whole-frame aligned and summed micrographs and used to create an initial raw particle stack after removing regions of the micrographs containing gold or large areas of aggregation. The raw particle stack was extracted using a box size of 256 pixels, and was immediately binned by 2 in reciprocal space, to a pixel size of 2.62 Å per pixel and a box size of 128 pixels. Initial 2D classification in Relion indicated a mixture of particle populations from sub-stoichiometric binding of sCD4 and 17b. Particles contributing to classes that did not produce clear structural features were discarded, leaving 98,845 particles. Next, to separate the different particle populations, this stack was subjected to competitive sorting using 3 initial models—trimer alone, trimer + sCD4, trimer + sCD4 + 17b Fab—in an identical manner as previously described37. This resulted in three separate stacks containing 69,105 particles (trimer alone), 6,342 particles (trimer + sCD4), and 23,398 particles (trimer + sCD4 + 17b). Each of these individual stacks was then subjected to 4-model classification in Relion, using 60 Å as the initial low-pass filter value36, followed by parameter conversion and final classification in Frealign38. For each subgroup, the number of classes in Frealign was varied from 2 to 4, and a final value was selected such that the classification would produce at least two nearly identical models, as described earlier39,40. Within the stack comprising the trimer alone or the trimer + sCD4, this procedure resulted in removal of bad particles that did not produce interpretable maps (Supplementary Fig. 5c); within the stack comprising the trimer + sCD4 + 17b, this resulted in one low resolution class that was discarded, as well as three classes that differed in the relative configuration of the 17b Fab, but that were otherwise very similar. Final resolutions (FSC 0.143) are around 10.5 Å (BG505CD4), and around 8.6 Å (BG505CD4–17b).
Model building and refinement
Model building and refinement steps followed those previously described41. In brief, initial homology models of B41 gp120 and gp41 were generated using Modeller42 and the crystal structure of ligand-free BG505 SOSIP.664 (PDB: 4ZMJ) as a template for B41LF and B41b12, while the crystal structure of gp120 bound to sCD4 and 17b (PDB: 1GC1) was used to model gp120 of B41CD4–17b. Coordinates for sCD4, 17b, b12, and PGV04 were obtained from PDB IDs 1GC1, 1RZ8, 2NY7, and 3J5M, respectively. Individual chains were fit into the respective cryo-EM maps using UCSF Chimera43 and refined using a combination of Rosetta density-guided iterative local refinement44, RosettaRelax45, and manual building in COOT46. Glycans were built using an idealized Man9 model that was placed into corresponding glycan density in UCSF Chimera. Torsion angles were adjusted in UCSF Chimera until good agreement was achieved between map and model. Sugar molecules with disordered or no density were removed. The final models were further refined using Phenix47 real space refinement without NCS constraints. Glycans were validated by pdb-care48 and CARP49 and structures were evaluated using EMRinger50 and Molprobity51. Figures were generated in UCSF Chimera or Pymol52. Buried surface area calculations were performed using PDBePISA53 and root-mean-square deviation (r.m.s.d.) analysis was performed in UCSF Chimera. Map-fitting cross correlations were calculated using the Fit In Map feature in UCSF Chimera.
Negative-stain electron microscopy
A stock of B41 SOSIP.664 was thawed on ice and 3 μl was adsorbed onto a Cu400 carbon-coated grid for 10 s before blotting, and then stained for 60 s with 2% (w/v) uranyl formate. The remaining stock was then incubated at 37 °C for 1 h on a heat block. Subsequently, 3 μl was removed and immediately adsorbed onto a carbon-coated grid followed by staining. The stock was again incubated on ice for 1 h before a third grid was prepared using the same method. Data were collected and processed as described in ref. 14.
Data availability
Cryo-EM reconstructions have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-8713, EMD-8714, EMD-8715, EMD-8716, EMD-8717, EMD-8729 and EMD-8730. Atomic models of B41b12 and B41CD4–17b have been deposited in the Protein Data Bank under accession numbers 5VN8 and 5VN3, respectively.
References
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008)
Lindemann, D., Steffen, I. & Pöhlmann, S. in Viral Entry into Host Cells (eds Pöhlmann, S. & Simmons, G. ) 128–149 (Springer New York, 2013)
Lee, J. H., Ozorowski, G. & Ward, A. B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016)
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013)
Burton, D. R. & Hangartner, L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 34, 635–659 (2016)
Burton, D. R. & Mascola, J. R. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 (2015)
Klein, F. et al. Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204 (2013)
West, A. P., Jr et al. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156, 633–648 (2014)
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008)
Tran, E. E. H. et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8, e1002797 (2012)
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998)
Guttman, M. et al. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22, 974–984 (2014)
Munro, J. B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014)
Pugach, P. et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J. Virol. 89, 3380–3395 (2015)
Sanders, R. W. et al. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 (2015)
Ringe, R. P. et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl Acad. Sci. USA 110, 18256–18261 (2013)
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015)
Finzi, A. et al. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37, 656–667 (2010)
Myszka, D. G. et al. Energetics of the HIV gp120–CD4 binding reaction. Proc. Natl Acad. Sci. USA 97, 9026–9031 (2000)
Pancera, M. et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl Acad. Sci. USA 107, 1166–1171 (2010)
Liu, Q. et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017)
Zhou, T. et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013)
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007)
Wang, J., Sen, J., Rong, L. & Caffrey, M. Role of the HIV gp120 conserved domain 1 in processing and viral entry. J. Biol. Chem. 283, 32644–32649 (2008)
Anastassopoulou, C. G. et al. Resistance of a human immunodeficiency virus type 1 isolate to a small molecule CCR5 inhibitor can involve sequence changes in both gp120 and gp41. Virology 413, 47–59 (2011)
Anastassopoulou, C. G., Ketas, T. J., Klasse, P. J. & Moore, J. P. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl Acad. Sci. USA 106, 5318–5323 (2009)
Tan, Q. et al. Structure of the CCR5 chemokine receptor–HIV entry inhibitor maraviroc complex. Science 341, 1387–1390 (2013)
Carragher, B. et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132, 33–45 (2000)
Potter, C. S. et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 77, 153–161 (1999)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009)
Ogura, T., Iwasaki, K. & Sato, C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J. Struct. Biol. 143, 185–200 (2003)
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013)
Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013)
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012)
Scheres, S. H. W. et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods 4, 27–29 (2007)
Kirchdoerfer, R. N. et al. Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 (2016)
Webb, B. & Sali, A. in Current Protocols in Bioinformatics (John Wiley & Sons, Inc., 2002)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015)
DiMaio, F., Tyka, M. D., Baker, M. L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using rosetta. J. Mol. Biol. 392, 181–190 (2009)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Lütteke, T. & von der Lieth, C.-W. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics 5, 69 (2004)
Lütteke, T., Frank, M. & von der Lieth, C.-W. Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 33, D242–D246 (2005)
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8 (2015)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Acknowledgements
This work was supported by the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1AI100663 (I.A.W., A.B.W.), NIH HIVRAD Grant P01 AI110657 (I.A.W., J.P.M., A.B.W.), NIH Grant R56 AI084817 (I.A.W.), and NIH Grant P50 GM103368 (D.L.). This is manuscript #29422 of The Scripps Research Institute.
Author information
Authors and Affiliations
Contributions
G.O., N.d.V., D.L., and A.B.W. designed the experiments. G.O., J.L.T., J.C., R.L.S., P.P., and A.C. produced the reagents. G.O. performed negative stain EM experiments. N.d.V., G.O., and J.P. prepared cryo-EM grids. N.d.V., J.P., G.O., and D.L. collected the electron microscopy data. D.L. processed the BG505 SOSIP data and J.P. processed the B41 SOSIP data. C.A.C. generated initial homology models and G.O. built and refined atomic models. G.O., J.P., and A.B.W. wrote the manuscript. G.O, J.P., N.d.V., J.P.M., I.A.W., and A.B.W. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks R.W. Doms, W. Mothes and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Cryo-EM statistics of B41CD4, B41CD4–17b and B41b12.
a, Fourier shell correlations. b, Local resolution estimates. c, Angular distribution plots. d, Fourier shell correlations for BG505CD4 and BG505CD4–17b.
Extended Data Figure 2 Cryo-EM reconstruction of ligand-free B41 SOSIP.664.
a, The ‘breathing’ of B41 SOSIP.664 trimers is suggested by negative-stain 2D class averages with representative open (blue) and closed (orange) phenotypes highlighted. Sample temperature influences the percentage of open and closed phenotypes, and the open state is reversible. b, Cryo-EM 2D class averages also suggest flexibility of the gp120 subunits relative to one another as evidenced by the blurring of gp120 subunits in the class averages. c, Cryo-EM reconstruction coloured by local resolution for B41LF. d, The segmented cryo-EM map coloured by component (gp120, gp41 and glycans). The top view is defined as looking towards the viral membrane. e, Fourier shell correlation (FSC) for B41LF. f, Superposition of the X-ray structure of ligand-free BG505 SOSIP.664 (PDB: 4ZMJ) onto a homology model of B41 SOSIP.664 refined into the cryo-EM map (Cα r.m.s.d., ~1.2 Å). All three protomers are presented in the top view (left), and only a single protomer is shown in the side view for clarity (middle). Example density corresponding to the V4 region and surrounding glycans (right).
Extended Data Figure 3 Comparison of B41CD4–17b models.
a, Fitting of the cryo-EM B41CD4–17b model into a cryo-electron tomography reconstruction of surface-expressed Env in complex with sCD4 and 17b. b, Alignment of gp120 in complex with CD4 and 17b from the cryo-EM and X-ray crystallography models (PDB: 1GC1). The Cα r.m.s.d. between our model and the X-ray structure of gp120 core in complex with sCD4 and 17b (PDB: 1GC1) is around 1.5 Å, which is relatively low considering only 79% identity between the BG505 and B41 Env sequences. c, Map fitting of B41CD4 into B41CD4–17b (low pass filtered to 5.2 Å) results in a cross correlation of 93%. d, Cartoon representation of the major changes involved in the formation of the α0 helix. Pre-fusion model based on PDB: 5CEZ.
Extended Data Figure 4 Conformational differences between pre-fusion, b12-bound and CD4-bound states.
a, Alignment of gp120 from the cryo-EM models of B41LF and B41CD4–17b illustrates a large displacement of the V1, V2, and V3 loops. b, Fitting of the B41b12 model into the map of B41CD4–17b reveals a steric barrier created by b12 that prevents the translocation of V1/V2, which would clash with the antibody heavy chain. c, Glycan N262 is repositioned away from the C1 domain of gp120 upon sCD4 binding as a result of V3 translocation (left). The relative positions of N262 in B41LF and B41CD4–17b are supported by continuous density (right). d, Comparison of the cryo-EM model of B41CD4–17b to cryo-EM reconstructions of BG505CD4 and BG505CD4–17b. e, Relative movements of variable loops between pre-fusion and CD4-bound states of B41 SOSIP (left). Loss of density for the V3 loop in both B41CD4 and B41CD4–17b maps suggests that V3 movement is in response to priming by CD4 and not caused by 17b or the co-receptor (right).
Extended Data Figure 5 Cryo-EM reconstruction of B41 SOSIP.664 in complex with PGV04 Fab.
a, Segmented cryo-EM map with components coloured according to the key. b, Docking of BG505 + PGV04 (PDB: 3J5M) and B41LF models into the B41 + PGV04 reconstruction. Comparison with BG505 bound to PGV04 demonstrates a high degree of structural similarity (91% correlation between the two cryo-EM maps), and docking of the B41LF model into the B41PGV04 map results in excellent agreement, with the B41LF backbone atoms falling into density as well as alignment of PNGS asparagine residues with glycan density (94% correlation between B41LF and B41PGV04 maps). c, FSC of B41PGV04. d, gp120 rotation and movement away from the central axis in the b12-bound state. e, Comparison of VRC01 and b12 epitopes with respect to the open, b12-bound conformation of B41 SOSIP.664. f, Docking of either the crystal structure of gp120–b12 or the B41b12 cryo-EM model into the ECT reconstruction of the HIV-1 strain BaL in complex with b12 (EMDB-5018) reveals differences in relative movement and rotation of gp120 from the trimer axis and ultimate position of b12.
Extended Data Figure 6 Comparison of the B41b12 cryo-EM model to the X-ray model of gp120 in complex with b12 Fab, and glycan repositioning.
a, Major contacts between b12 Fab and gp120 highlighted based on the cryo-EM model. Sequence alignment between BG505 and B41 Env for a segment of V2, with N-linked glycans coloured green. b, Alignment of gp120 in complex with b12 from the cryo-EM and X-ray crystallography models (PDB: 2NYZ) (left). The crystal structure of b12 Fab docked into the B41b12 cryo-EM map, revealing that the elbow angle is preserved (middle). When aligned to gp120, the X-ray model reveals a slightly different b12 angle of approach with respect to the cryo-EM model (right). These differences may arise owing to a more stable, neutralization-resistant tier-2 virus (B41) versus a more flexible laboratory-adapted tier-1 BaL pseudovirus, the stabilizing SOSIP modifications used in the soluble constructs (although none are located at the CD4bs epitope or gp120 core), or the low resolution of the cryoET map. c, The pre-fusion arrangement of gp120 does not allow for b12 binding to B41 SOSIP.664 owing to clashes between the framework regions of the antibody and portions of a neighbouring gp120 monomer (V3 and glycans). d, Rotation of the N197 glycan requires the gp120 subunits to open up and move away from one another. e, f, Glycan N197 in the b12 epitope acts as a steric barrier in the pre-fusion state and rearranges and moves away from the b12 epitope to allow for b12 binding.
Extended Data Figure 7 Cryo-EM density of various stabilizing interactions in B41CD4–17b.
a, b, Stereo images of cryo-EM density of specified contour levels for K574–D107 (a), and the α0 HR1 cap of B41CD4–17b (b). c, d, Fusion peptide and fusion peptide proximal region electron density for B41b12 (c) and B41CD4–17b (d). e, Stereo image of pocket protecting the fusion peptide in the CD4-bound state. f, Stereo image of electron density of the Trp clasp region in B41CD4–17b. g, HR1 three-helix bundle rearrangement between pre-fusion and CD4-bound states. h, gp41 arrangement in B41CD4–17b. The fusion peptide proximal region and HR1N pack against regions of HR2 from two different protomers.
Extended Data Figure 8 Overview of HIV-1 Env conformational states.
Various biophysical data strongly suggest that the pre-fusion trimer is in equilibrium of reversible open and closed states. b12 recognizes a more open state and traps the trimer in an irreversible intermediate state that can no longer have a role in host cell fusion. CD4, on the other hand, induces a stable, fusion intermediate that displays the co-receptor binding site and primes the fusion peptide to move to a more centralized location in the trimer interface. It is only after binding of CXCR4/CCR5 to CD4-bound Env that additional fusion steps occur, highlighted by the full formation of a three-helix bundle before final condensation into a six-helix bundle. When a dimer of CCR5 from a crystal structure is docked on top of the trimer structure, the N termini of the two co-receptors are situated proximal to the co-receptor binding sites on gp120.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion and Supplementary References. (PDF 158 kb)
Conformational changes between B41LF and B41b12 or B41CD4/17b.
Morph between Cα coordinates of ligand-free and either b12-bound or CD4/17b-bound B41 SOSIP.664. CryoEM maps of each complex are shown for reference. Morphing and video rendering were done using UCSF Chimera. (MOV 18167 kb)
CD4-induced allosteric network in B41CD4/17b.
Morph between ligand-free and CD4/17b-bound B41 SOSIP.664, highlighting residues described in figure 2a. CryoEM maps of each complex are shown at the beginning and end of video for reference. Morphing and video rendering were done using UCSF Chimera. (MOV 29150 kb)
Formation of helix α0 induced by CD4 binding
Morph between ligand-free and CD4/17b-bound B41 SOSIP.664, highlighting the formation of α0 helix (green). CryoEM maps of each complex are shown at the beginning and end of video for reference. Morphing and video rendering were done using UCSF Chimera. (MOV 28733 kb)
Formation of a new pocket that sequesters the fusion peptide in B41CD4/17b and B41b12.
Morph between ligand-free and CD4/17b-bound B41 SOSIP.664, highlighting the movement of the fusion peptide from the solvent to a newly-formed pocket (green). Coordinates for the ligand-free state fusion peptide were taken from PDB ID 5CEZ. CryoEM maps of each complex are shown at the beginning and end of video for reference. Morphing and video rendering were done using UCSF Chimera. (MOV 29383 kb)
Rights and permissions
About this article
Cite this article
Ozorowski, G., Pallesen, J., de Val, N. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017). https://doi.org/10.1038/nature23010
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23010
This article is cited by
-
Conformational antigenic heterogeneity as a cause of the persistent fraction in HIV-1 neutralization
Retrovirology (2023)
-
HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes
Nature (2023)
-
Assessing immunogenicity barriers of the HIV-1 envelope trimer
npj Vaccines (2023)
-
Intermediate conformations of CD4-bound HIV-1 Env heterotrimers
Nature (2023)
-
Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.