Abstract
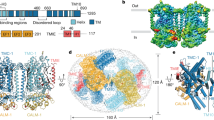
Mechanosensory transduction for senses such as proprioception, touch, balance, acceleration, hearing and pain relies on mechanotransduction channels, which convert mechanical stimuli into electrical signals in specialized sensory cells1. How force gates mechanotransduction channels is a central question in the field, for which there are two major models. One is the membrane-tension model: force applied to the membrane generates a change in membrane tension that is sufficient to gate the channel, as in the bacterial MscL channel and certain eukaryotic potassium channels2,3,4,5. The other is the tether model: force is transmitted via a tether to gate the channel. The transient receptor potential (TRP) channel NOMPC is important for mechanosensation-related behaviours such as locomotion, touch and sound sensation across different species including Caenorhabditis elegans6, Drosophila7,8,9 and zebrafish10. NOMPC is the founding member of the TRPN subfamily11, and is thought to be gated by tethering of its ankyrin repeat domain to microtubules of the cytoskeleton12,13,14,15. Thus, a goal of studying NOMPC is to reveal the underlying mechanism of force-induced gating, which could serve as a paradigm of the tether model. NOMPC fulfils all the criteria that apply to mechanotransduction channels1,7 and has 29 ankyrin repeats, the largest number among TRP channels. A key question is how the long ankyrin repeat domain is organized as a tether that can trigger channel gating. Here we present a de novo atomic structure of Drosophila NOMPC determined by single-particle electron cryo-microscopy. Structural analysis suggests that the ankyrin repeat domain of NOMPC resembles a helical spring, suggesting its role of linking mechanical displacement of the cytoskeleton to the opening of the channel. The NOMPC architecture underscores the basis of translating mechanical force into an electrical signal within a cell.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Árnadóttir, J. & Chalfie, M. Eukaryotic mechanosensitive channels. Annu. Rev. Biophys. 39, 111–137 (2010)
Anishkin, A. & Kung, C. Stiffened lipid platforms at molecular force foci. Proc. Natl Acad. Sci. USA 110, 4886–4892 (2013)
Brohawn, S. G., del Mármol, J. & MacKinnon, R. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science 335, 436–441 (2012)
Brohawn, S. G., Campbell, E. B. & MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014)
Lolicato, M., Riegelhaupt, P. M., Arrigoni, C., Clark, K. A. & Minor, D. L., Jr. Transmembrane helix straightening and buckling underlies activation of mechanosensitive and thermosensitive K2P channels. Neuron 84, 1198–1212 (2014)
Li, W., Feng, Z., Sternberg, P. W. & Xu, X. Z. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature 440, 684–687 (2006)
Yan, Z. et al. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature 493, 221–225 (2013)
Effertz, T., Wiek, R. & Göpfert, M. C. NompC TRP channel is essential for Drosophila sound receptor function. Curr. Biol. 21, 592–597 (2011)
Cheng, L. E., Song, W., Looger, L. L., Jan, L. Y. & Jan, Y. N. The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron 67, 373–380 (2010)
Sidi, S., Friedrich, R. W. & Nicolson, T. NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science 301, 96–99 (2003)
Walker, R. G., Willingham, A. T. & Zuker, C. S. A. A Drosophila mechanosensory transduction channel. Science 287, 2229–2234 (2000)
Zhang, W. et al. Ankyrin repeats convey force to gate the NOMPC mechanotransduction channel. Cell 162, 1391–1403 (2015)
Liang, X. et al. A NOMPC-dependent membrane-microtubule connector is a candidate for the gating spring in fly mechanoreceptors. Curr. Biol. 23, 755–763 (2013)
Lee, G. et al. Nanospring behaviour of ankyrin repeats. Nature 440, 246–249 (2006)
Howard, J. & Bechstedt, S. Hypothesis: a helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr. Biol. 14, R224–R226 (2004)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016)
Paulsen, C. E., Armache, J. P., Gao, Y., Cheng, Y. & Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517 (2015)
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007)
Ahuja, S . et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science 350, aac5464 (2015)
Wu, J. et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature 537, 191–196 (2016)
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013)
Saotome, K., Singh, A. K., Yelshanskaya, M. V. & Sobolevsky, A. I. Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511 (2016)
Zubcevic, L. et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 23, 180–186 (2016)
Huynh, K. W. et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 7, 11130 (2016)
Venkatachalam, K. & Montell, C. TRP channels. Annu. Rev. Biochem. 76, 387–417 (2007)
Lin, Z. et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am. J. Hum. Genet. 90, 558–564 (2012)
Teng, J., Loukin, S. H., Anishkin, A. & Kung, C. L596-W733 bond between the start of the S4–S5 linker and the TRP box stabilizes the closed state of TRPV4 channel. Proc. Natl Acad. Sci. USA 112, 3386–3391 (2015)
Wang, C. et al. Structural basis of diverse membrane target recognitions by ankyrins. eLife 3, 1–22 (2014)
Cox, C. D. et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 7, 10366 (2016)
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014)
Fridy, P. C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014)
Booth, D. S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 58, e3227 (2011)
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Elmlund, H., Elmlund, D. & Bengio, S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure 21, 1299–1306 (2013)
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015)
Chovancova, E. et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput. Biol. 8, e1002708 (2012)
McKenney, R. J., Huynh, W., Tanenbaum, M. E., Bhabha, G. & Vale, R. D. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 (2014)
Schroeder, C. M. & Vale, R. D. Assembly and activation of dynein–dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214, 309–318 (2016)
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using μmanager. Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010)
Acknowledgements
We thank R. Mackinnon, T. Xiao and W. Wang for advice on protein purification, S. Gründer for sharing construct, and members from laboratories of D. Minor, D. Julius and R. Vale for sharing equipment and reagents. We also thank colleagues in our laboratories for discussions, M. Braunfeld and C. Kennedy for technical support, and Z. Yu and his colleagues at the HHMI Janelia Cryo-EM Facility for help with data acquisition. This work was supported by grants from the NIH (R01NS069229 to L.Y.J., 5R37NS040929, 1R35NS097227 to Y.-N.J., R01GM098672 and S10OD020054 to Y.C.), and by a UCSF Program for Breakthrough Biomedical Research (Y.C.). L.Y.J., Y.-N.J. and Y.C. are Investigators with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
P.J. and D.B. designed and performed most biochemical and cryo-EM experiments. Y.G. performed electrophysiology experiments, and W.Z. performed mutagenesis and initial electrophysiology studies of NOMPC. Z.G. assisted with biochemical experiments. W.H. performed microtubule-binding assay. S.W. helped with cryo-EM experiments. S.M. performed surface staining, T.C. performed mutagenesis. L.Y.J., Y.-N.J. and Y.C supervised experiments and data analysis. P.J., D.B., L.Y.J., Y.-N.J. and Y.C. wrote the manuscript. All authors contributed to manuscript preparations.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks E. McCleskey, K. Vinothkumar and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Verification of recombinant NOMPC activity.
a, Pressure-induced mechanogated current measured at −60 mV from outside-out patches excised from HEK293 cells untransfected (left) or transfected (right) with the NOMPC construct used for structure determination. b, TIRF microscopy images of surface anchored and fluorescently labelled microtubules (pseudo-coloured in magenta) are not decorated by purified strep-GFP alone (top) but by strep-GFP-tagged NOMPC (bottom), demonstrating that purified NOMPC is capable of binding microtubules, and that this binding is not through interaction between the Strep tag and residual streptavidin on the surface.
Extended Data Figure 2 Negative-stain analysis of purified NOMPC.
a, Size-exclusion chromatography of NOMPC after exchange from DDM into amphipol A8-35. The peak fraction corresponding to NOMPC tetramer (indicated by arrow) was examined by SDS–polyacrylamide gel electrophoresis. The NOMPC monomer (approximately 190 kDa) band is indicated by an arrowhead. The top bands may correspond to incompletely disassociated NOMPC oligomers. b, Size-exclusion chromatography of NOMPC reconstituted into lipid nanodisc with MSP2N2. The peak fraction corresponding to NOMPC is indicated by arrow. c, d, Raw micrographs of NOMPC–amphipol (c) and NOMPC–nanodisc (d) samples examined by negative-stain EM. Both showed mono-dispersed and homogeneous particles. e, f, 2D class averages of NOMPC particles in amphipol (e) or nanodisc (f) by negative-stain EM, with the nanodisc sample showing better ordered features than the amphipol sample.
Extended Data Figure 3 Cryo-EM analysis of NOMPC in amphipol.
a, A raw cryo-EM micrograph of NOMPC recorded as described in Methods. b, 2D class averages of the cryo-EM micrographs with a particle box size of 400 pixels (486 Å). c, Euler angle distribution of all particles used for the final 3D reconstruction. The size of the sphere is proportional to the amount of particles visualized from that specific orientation. d, Final unsharpened 3D density map of NOMPC-amphipol colored with local resolution at a low isosurface level (top left) to enable visualization of the amphipol layer, and at a high isosurface level in side (top right), top (bottom right) and bottom (bottom left) views. e, FSC curves between two independently refined half maps before (red) and after (blue) post-processing in RELION, indicated with resolutions corresponding to FSC = 0.143. f, Density maps of NOMPC in amphipol (top) and in nanodisc (bottom) with a C1 symmetry derived from all particles picked. Comparison of the maps shows NOMPC-nanodisc is closer to a four-fold symmetric structure.
Extended Data Figure 4 Cryo-EM analysis of NOMPC reconstituted in nanodisc.
a, A raw cryo-EM micrograph of NOMPC recorded as described in Methods. b, 2D class averages of the cryo-EM micrographs with a particle box size of 400 pixels (486 Å). c, Euler angle distribution of all particles used for the final 3D reconstruction. The size of the sphere is proportional to the amount of particles visualized from that specific orientation. d, Slices through the unsharpened density map at different levels along the channel symmetry axis. The slice numbers starting from the cytoplasmic side are marked. e, Final unsharpened 3D density map of NOMPC–nanodisc coloured with local resolution at a low isosurface level (top left) to enable visualization of the lipid bilayer, and at a high isosurface level in side (top right), top (bottom right) and bottom (bottom left) views. f, FSC curves between two independently refined half maps before (red) and after (blue) post-processing in RELION, indicated with resolutions corresponding to FSC = 0.143. g, Cross-validation using FSC curves of the density map calculated from the refined model versus half map 1 (work, green), versus half map 2 (free, pink) and versus summed map (blue).
Extended Data Figure 5 Selected segments of cryo-EM density.
a–j, Representative cryo-EM densities of various NOMPC domains as indicated are superimposed on the atomic model. k, Density map of pre-S1 elbow, S1 and S4. l, Density map of stacked S4–S5 linker, TRP domain and linker helices that couple ARs to the pore through domain interactions. All the density maps (a–l) are shown as cyan meshes, and the model is shown as sticks and coloured according to atom type (C: light grey; N: blue; O: red; S: yellow).
Extended Data Figure 6 Atomic model of NOMPC.
a–d, Ribbon diagrams of NOMPC atomic model for residues Asn 125–Trp 1602 and Gly1689–Arg1670. The entire AR domain of 29 ARs was resolved. e, Ribbon diagrams showing two views of one NOMPC subunit denoting specific domains.
Extended Data Figure 7 Pore profile and channel properties of NOMPC and NOMPC mutants.
a, Pore radius calculated by the CAVER program. b, I–V curves from the steady-state currents of whole-cell recording from S2 cells expressing NOMPC or NOMPC mutants suggesting that both W1572A and I1554A mutations resulted in large basal current in the absence of applied pressure (wild type: n = 7; W1572A: n = 7; I1554A: n = 3). Cells expressing I1554A mutant channels displayed very large basal currents and were not amenable to recording from excised patch recording of mechanogated current. c, Dose-dependent curves of pressure-induced mechanogated currents measured at −60 mV from outside-out patches excised from S2 cells expressing NOMPC or NOMPC mutants (n = 7). d, Representative images of unpermeabilized staining of S2 cells expressing control proteins or His1423Ala mutant. The surface staining signal from approximately 35 cells were visually surveyed under fluorescent microscope. All cells expressing wild-type (middle) and His1423Ala mutant (bottom) NOMPC showed similar levels of surface expression, indicating that the His1423Ala mutant is properly localized to the plasma membrane like the wild type. All error bars (b, c) denote s.d.
Extended Data Figure 8 3D classification of NOMPC-nanodisc particles.
The flowchart of classification procedures with RELION is shown. After 1 round of 2D classification, 190,879 particles were subjected to 3D refinement, yielding a 3.7 Å map (C4 imposed). Following 3D classification of these particles, 175,314 particles were selected and refined, yielding the final map at 3.55 Å resolution (C4 imposed), which was used to build the ‘consensus model’. Further classification of these 175,314 particles using a mask to exclude all regions outside the AR domain gave 3 major classes, which were subsequently refined to around 3.8–4.0 Å resolution.
Extended Data Figure 9 Flexibility of AR domains and model of NOMPC mechanogating.
a, Superposition of NOMPC AR domains from three classes are shown in three views as indicated. Overall, the entire AR domains from all three classes overlap well with each other. There are small differences in some ARs, which are caused by a slight shift of individual ARs as a rigid body, suggesting small mobility and plasticity of the AR domain. b, Superposition of NOMPC AR domain (blue) with human ankyrin-B AR domain (pink) shown in three views as indicated, suggesting that elastic deformation of NOMPC AR domain under pressure could potentially be more notable than the shift presumably caused by thermal motion as shown in a. A peptide from the C-terminal region of ankyrin-R, which was added to stabilize ankyrin-B ARs for crystallization by forming an auto-inhibitory segment (AS) structure, is also shown (AnkR AS, in orange). c, Schematic of NOMPC (without precise depiction of domain swap between neighbouring subunits) showing the N terminus tethered to a microtubule. Mechanical force is transduced from the microtubule cytoskeleton to NOMPC, possibly causing lateral movement, extension, compression or torsion of the AR domain. d, e, Movement of the AR domain that immediately precedes the linker helices results in displacement of the TRP domain and S4–S5 linker that are connected to the ends of the pore domain, triggering channel opening.
Extended Data Figure 10 Sequence alignment of NOMPC orthologues.
Sequence homology of NOMPC orthologues were analysed by clustal omega. The conserved residues are highlighted. The two residues (His1423 and Trp1572), which were shown in this study to be crucial for mechanogating, are marked by red triangles. Secondary structure elements are indicated above the sequence.
Supplementary information
Supplementary Information
This file contains a supplementary discussion and a table of the summary of cryo-EM data collection and model refinement.
Supplementary Figure
This file contains the uncropped SDS PAGE gel for extended data figure 2a.
Class 1 to class 2 transition
Animated video shows morph between atomic models generated from “class 1” and “class 2” maps (Ext. Data Fig. 7). Atomic models were generated by real space refinement of the consensus model into the maps for the corresponding classes in PHENIX.
Class 1 to class 3 transition
Animated video shows morph between atomic models generated from “class 1” and “class 3” maps (Ext. Data Fig. 7). Atomic models were generated as in Extended Data Video 1.
Class 2 to class 3 transition
Animated video shows morph between atomic models generated from “class 2” and “class 3” maps (Ext. Data Fig. 7). Atomic models were generated as in Extended Data Video 1.
Rights and permissions
About this article
Cite this article
Jin, P., Bulkley, D., Guo, Y. et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017). https://doi.org/10.1038/nature22981
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22981
This article is cited by
-
Nuclear pore protein POM121 regulates subcellular localization and transcriptional activity of PPARγ
Cell Death & Disease (2024)
-
Piezo1 channel exaggerates ferroptosis of nucleus pulposus cells by mediating mechanical stress-induced iron influx
Bone Research (2024)
-
Ultrafast underwater self-healing piezo-ionic elastomer via dynamic hydrophobic-hydrolytic domains
Nature Communications (2024)
-
Molecular architecture and gating mechanisms of the Drosophila TRPA1 channel
Cell Discovery (2023)
-
Cryo-EM structure of TMEM63C suggests it functions as a monomer
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.