Abstract
Glucagon-like peptide 1 (GLP-1) regulates glucose homeostasis through the control of insulin release from the pancreas. GLP-1 peptide agonists are efficacious drugs for the treatment of diabetes. To gain insight into the molecular mechanism of action of GLP-1 peptides, here we report the crystal structure of the full-length GLP-1 receptor bound to a truncated peptide agonist. The peptide agonist retains an α-helical conformation as it sits deep within the receptor-binding pocket. The arrangement of the transmembrane helices reveals hallmarks of an active conformation similar to that observed in class A receptors. Guided by this structural information, we design peptide agonists with potent in vivo activity in a mouse model of diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Todd, J. F. & Bloom, S. R. Incretins and other peptides in the treatment of diabetes. Diabet. Med. 24, 223–232 (2007)
Chatterjee, S., Ghosal, S. & Chatterjee, S. Glucagon-like peptide-1 receptor agonists favorably address all components of metabolic syndrome. World J. Diabetes 7, 441–448 (2016)
Mentlein, R., Gallwitz, B. & Schmidt, W. E. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7-36)amide, peptide histidine methionine and is responsible for their degradation in human serum. Eur. J. Biochem. 214, 829–835 (1993)
Deacon, C. F., Johnsen, A. H. & Holst, J. J. Degradation of glucagon-like peptide-1 by human plasma in vitro yields an N-terminally truncated peptide that is a major endogenous metabolite in vivo. J. Clin. Endocrinol. Metab. 80, 952–957 (1995)
Willard, F. S., Bueno, A. B. & Sloop, K. W. Small molecule drug discovery at the glucagon-like peptide-1 receptor. Exp. Diabetes Res. 2012, 709893 (2012)
Mann, R. et al. Peptide binding at the GLP-1 receptor. Biochem. Soc. Trans. 35, 713–716 (2007)
Underwood, C. R. et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 285, 723–730 (2010)
Runge, S., Thøgersen, H., Madsen, K., Lau, J. & Rudolph, R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 283, 11340–11347 (2008)
Siu, F. Y. et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449 (2013)
Jazayeri, A. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–277 (2016)
Haque, T. S. et al. Identification of potent 11mer glucagon-like peptide-1 receptor agonist peptides with novel C-terminal amino acids: homohomophenylalanine analogs. Peptides 31, 950–955 (2010)
Haque, T. S. et al. Exploration of structure-activity relationships at the two C-terminal residues of potent 11mer glucagon-like peptide-1 receptor agonist peptides via parallel synthesis. Peptides 31, 1353–1360 (2010)
Mapelli, C. et al. Eleven amino acid glucagon-like peptide-1 receptor agonists with antidiabetic activity. J. Med. Chem. 52, 7788–7799 (2009)
Serrano-Vega, M. J., Magnani, F., Shibata, Y. & Tate, C. G. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc. Natl Acad. Sci. USA 105, 877–882 (2008)
Lebon, G. et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474, 521–525 (2011)
White, J. F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012)
Yang, L. et al. Conformational states of the full-length glucagon receptor. Nat. Commun. 6, 7859 (2015)
Hollenstein, K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013)
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA 110, 5211–5216 (2013)
Tom Burnley, B ., Afonine, P. V ., Adams, P. D. & Gros, P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife 2012, 1–29 (2012)
Yang, D. et al. Structural determinants of binding the seven-transmembrane domain of the glucagon-like peptide-1 receptor (GLP-1R). J. Biol. Chem. 291, 12991–13004 (2016)
Dods, R. L. & Donnelly, D. The peptide agonist-binding site of the glucagon-like peptide-1 (GLP-1) receptor based on site-directed mutagenesis and knowledge-based modelling. Biosci. Rep. 36, e00285 (2015)
Wootten, D. et al. The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell 165, 1632–1643 (2016)
Coopman, K. et al. Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor. Mol. Endocrinol. 25, 1804–1818 (2011)
Xiao, Q., Jeng, W. & Wheeler, M. B. Characterization of glucagon-like peptide-1 receptor-binding determinants. J. Mol. Endocrinol. 25, 321–335 (2000)
Koole, C. et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J. Biol. Chem. 287, 3642–3658 (2012)
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880 (2002)
Moon, M. J. et al. Ligand binding pocket formed by evolutionarily conserved residues in the glucagon-like peptide-1 (GLP-1) receptor core domain. J. Biol. Chem. 290, 5696–5706 (2015)
López de Maturana, R., Treece-Birch, J., Abidi, F., Findlay, J. B. C. & Donnelly, D. Met-204 and Tyr-205 are together important for binding GLP-1 receptor agonists but not their N-terminally truncated analogues. Protein Pept. Lett. 11, 15–22 (2004)
Koth, C. M. et al. Molecular basis for negative regulation of the glucagon receptor. Proc. Natl Acad. Sci. USA 109, 14393–14398 (2012)
Mason, J. S. et al. High end GPCR design: crafted ligand design and druggability analysis using protein structure, lipophilic hotspots and explicit water networks. In Silico Pharmacol. 1, 23 (2013)
Goodford, P. J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28, 849–857 (1985)
Sciabola, S. et al. High-throughput virtual screening of proteins using GRID molecular interaction fields. J. Chem. Inf. Model. 50, 155–169 (2010)
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011)
Deupi, X. & Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011)
Tehan, B. G., Bortolato, A., Blaney, F. E., Weir, M. P. & Mason, J. S. Unifying family A GPCR theories of activation. Pharmacol. Ther. 143, 51–60 (2014)
Wheeler, M. B. et al. Functional expression of the rat glucagon-like peptide-I receptor, evidence for coupling to both adenylyl cyclase and phospholipase-C. Endocrinology 133, 57–62 (1993)
Vilsbøll, T., Agersø, H., Krarup, T. & Holst, J. J. Similar elimination rates of glucagon-like peptide-1 in obese type 2 diabetic patients and healthy subjects. J. Clin. Endocrinol. Metab. 88, 220–224 (2003)
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011)
Nicholls, R. A., Long, F. & Murshudov, G. N. Low-resolution refinement tools in REFMAC5. Acta Crystallogr. D Biol. Crystallogr. 68, 404–417 (2012)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010)
Diederichs. K. & Karplus, P. A. Better models by discarding data? Acta Crystallogr D Biol Crystallogr. 69, 1215–1222 (2013)
Acknowledgements
We thank various colleagues past and present who have helped with the project. In particular we would like to acknowledge the contribution of K. Hollenstein, M. Koglin and C. Larner. We thank G. Brown for his help with coordinating peptide synthesis and radio-labelling and C. Scully for his assistance with the GRID analysis. We are grateful to R. Owen, J. Waterman and D. Axford at I24, Diamond Light Source, Oxford, UK for technical support.
Author information
Authors and Affiliations
Contributions
J.K., N.J.R. and A.J. carried out the conformational thermostabilization of constructs and determined the stability of the StaR in a panel of reagents/additives. A.H.B. and I.T. carried out the in vitro pharmacology. A.J.H.B. managed the in vivo studies. M.C. and S.P.A. designed the novel peptides, aided by A.B. and J.S.M. who designed the homology models and carried out in silico analyses of peptide binding. M.R. and J.C.E. designed the crystallization construct, and with C.F.V. performed and optimized protein expression and purification. M.R. and C.F.V. performed and optimized protein crystallization. M.R. and A.S.D. harvested crystals, collected and processed X-ray diffraction data, and solved and refined the structure. Project management was carried out by A.J., R.M.C., F.H.M. and M.W. The manuscript was prepared by M.R., A.J., A.S.D., A.J.H.B., M.C., R.M.C. and F.H.M. All authors contributed to the final editing and approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of Heptares Therapeutics Ltd and are shareholders in Sosei Group Corporation.
Additional information
Reviewer Information Nature thanks T. Schwartz, C. Siebold and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
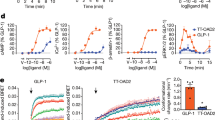
Extended Data Figure 1 Peptide 5 and in vitro pharmacology of wild-type GLP-1R.
a, Two-dimensional chemical plot of peptide 5 used in this study. b, Pharmacological characterization of 3H-peptide 1 at the wild-type GLP-1R. Affinity of peptide 1 for wild-type GLP-1R construct was measured using homologous competition experiments against three different concentration of 3H-peptide 1. cpm, counts per minute. Data are representative of four independent experiments and the Kd values are calculated as the arithmetic mean and s.e.m. c, Heterologous competition binding of exendin-4 and exendin-3 at the wild-type GLP-1R determined using 3H-peptide 1. Affinity constants (Ki) were calculated from IC50 values, using the Cheng–Prusoff equation and results are given as the arithmetic mean ± s.e.m.
Extended Data Figure 2 Stability, pharmacological characterization and functional activity of GLP-1R StaR.
a, Thermal stability (Tm) comparison of GLP-1R wild-type and StaR (containing the following point mutations: T207E, Q211A, D215R, L232F, G295A, T298A, C329A, P358A, G361A, H363V and V405A). Thermal stability was measured following solubilization in n-dodecyl-β-d-maltopyranoside supplemented with cholesteryl hemisuccinate (see Methods). Data representative of two independent experiments with Tm values calculated as the arithmetic mean and the standard deviation of the mean. b, Pharmacological characterization of GLP-1R wild-type and StaR. Affinity of peptide 1 for wild-type and StaR constructs was measured using homologous competition experiments against three different concentrations of 3H-peptide 1. Data representative of three independent experiments and the Kd values calculated as the arithmetic mean and the standard deviation of the mean. The difference in the means is not statistically significant as analysed by two-tailed t-test. c, d, cAMP response of GLP-1R wild-type and StaR in the presence of the indicated peptide agonists. e, Reported pEC50 values for each peptide agonist. Data presented is the arithmetic mean of three independent experiments. Error bars represent s.e.m. P values are calculated by multi-parametric two-way ANOVA.
Extended Data Figure 3 The GLP-1R StaR B-factors and electron density.
a, b, B-factor putty representation of the GLP-1R–peptide-5 structure (rainbow spectrum, blue to red = lowest to highest B-factors). c, d, representative 2Fo − Fc electron density contoured at 1.0σ across the orthosteric peptide-binding pocket of GLP-1R.
Extended Data Figure 4 Walkthrough of peptide 5 interactions with GLP-1R StaR.
a–k, Views moving from the N to C terminus of peptide 5. Four overlayed models generated by PHENIX ensemble refinement are shown in each panel to demonstrate the confidence that can be assigned to interactions with the receptor described in this study.
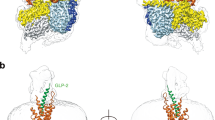
Extended Data Figure 5 Structural superposition of the GLP-1R peptide 5 crystal structure with the crystal structure of the isolated GLP-1R extracellular domain in complex with the GLP-1 peptide.
a, Tilted view from membrane of the GLP-1R represented as cartoon (cyan) with the peptide 5 agonist in stick representation and carbon, nitrogen and oxygen atoms coloured yellow, blue and red respectively; the ECD solved in isolation from the TMD of GLP-1R in cartoon representation is coloured orange, with the GLP-1 peptide coloured magenta. The superposition was achieved using equivalent residues from peptide 5 and the GLP-1 peptide. b, Rotation of the superposed structures in a to view from extracellular space, the relative difference in orientation of the ECD is denoted.
Extended Data Figure 6 Evaluation of the lipophilic hotspots on GLP-1R and interactions with peptide 5.
a, GRID hotspot analysis of the binding mode of peptide 5 showing the overlap of the Cap1, X2 and X3 groups of peptide 5 with lipophilic regions of the GLP-1R. GLP-1R is represented as cartoon (cyan) with the ECD coloured brown. The peptide 5 agonist is shown in stick representation with carbon, nitrogen and oxygen atoms coloured yellow, blue and red respectively. The c1 = GRID map is represented as mesh (orange) and contoured at −2.5 kcal mol−1. b, View as in a rotated by 180°.
Extended Data Figure 7 In vitro pharmacological and pharmacokinetic profiles of selected peptides.
a–d, Insulinotropic activities of GLP-1 and selected peptides on isolated rat pancreatic islets. Results are presented as mean ± s.e.m. (n = 6 each group) and analysed using a one-way analysis of variance and Dunnett’s post hoc test. Significant differences from basal responses are indicated with an asterisk (*P < 0.05). e, Pharmacokinetics of peptide 2 (1 mg kg−1), peptide 5 (1 mg kg−1) and peptide 8 (0.5 mg kg−1) following intravenous administration in male Sprague Dawley rats. Results presented as mean ± s.e.m. (n = 3). f, Pharmacokinetics of peptide 8 following subcutaneous administration in male CD1 mice. Results presented as mean ± s.e.m. (n = 3).
Supplementary information
Supplementary information
This file contains Supplementary Text and Data, 2 Supplementary Tables and Supplementary references. (PDF 413 kb)
Rights and permissions
About this article
Cite this article
Jazayeri, A., Rappas, M., Brown, A. et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546, 254–258 (2017). https://doi.org/10.1038/nature22800
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22800
This article is cited by
-
Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with Gs proteins
Cell Discovery (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Applications and prospects of cryo-EM in drug discovery
Military Medical Research (2023)
-
GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
Nature Communications (2023)
-
Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.