Abstract
A variety of tissue lineages can be differentiated from pluripotent stem cells by mimicking embryonic development through stepwise exposure to morphogens, or by conversion of one differentiated cell type into another by enforced expression of master transcription factors. Here, to yield functional human haematopoietic stem cells, we perform morphogen-directed differentiation of human pluripotent stem cells into haemogenic endothelium followed by screening of 26 candidate haematopoietic stem-cell-specifying transcription factors for their capacity to promote multi-lineage haematopoietic engraftment in mouse hosts. We recover seven transcription factors (ERG, HOXA5, HOXA9, HOXA10, LCOR, RUNX1 and SPI1) that are sufficient to convert haemogenic endothelium into haematopoietic stem and progenitor cells that engraft myeloid, B and T cells in primary and secondary mouse recipients. Our combined approach of morphogen-driven differentiation and transcription-factor-mediated cell fate conversion produces haematopoietic stem and progenitor cells from pluripotent stem cells and holds promise for modelling haematopoietic disease in humanized mice and for therapeutic strategies in genetic blood disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014)
Riddell, J. et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157, 549–564 (2014)
Sandler, V. M. et al. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511, 312–318 (2014)
Pereira, C. F. et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 13, 205–218 (2013)
Szabo, E. et al. Direct conversion of human fibroblasts to sub-lethal blood progenitors. Nature 468, 521–526 (2010)
Batta, K., Florkowska, M., Kouskoff, V. & Lacaud, G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 9, 1871–1884 (2014)
Doulatov, S. et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13, 459–470 (2013)
Dieterlen-Lievre, F. On the origin of haemopoietic stem cells in the avian embryo: an experimental approach. J. Embryol. Exp. Morphol. 33, 607–619 (1975)
Ivanovs, A. et al. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208, 2417–2427 (2011)
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010)
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010)
Dzierzak, E. & Speck, N. A. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008)
Kennedy, M. et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2, 1722–1735 (2012)
Ditadi, A. et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 17, 580–591 (2015)
Elcheva, I. et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 5, 4372 (2014)
Ivanova, N. B. et al. A stem cell molecular signature. Science 298, 601–604 (2002)
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11, 585–593 (2010)
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011)
Shultz, L. D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005)
Ferrell, P. I., Xi, J., Ma, C., Adlakha, M. & Kaufman, D. S. The RUNX1 +24 enhancer and P1 promoter identify a unique subpopulation of hematopoietic progenitor cells derived from human pluripotent stem cells. Stem Cells 33, 1130–1141 (2015)
Ditadi, A. & Sturgeon, C. M. Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods 101, 65–72 (2016)
McIntosh, B. E. et al. Nonirradiated NOD,B6.SCID Il2rγ−/−KitW41/W41 (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Rep. 4, 171–180 (2015)
Casero, D. et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 16, 1282–1291 (2015)
Notta, F . et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016)
Glodek, A. M. et al. Focal adhesion kinase is required for CXCL12-induced chemotactic and pro-adhesive responses in hematopoietic precursor cells. Leukemia 21, 1723–1732 (2007)
Hartz, B. et al. α4 integrin levels on mobilized peripheral blood stem cells predict rapidity of engraftment in patients receiving autologous stem cell transplantation. Blood 118, 2362–2365 (2011)
Smith-Berdan, S. et al. Robo4 cooperates with CXCR4 to specify hematopoietic stem cell localization to bone marrow niches. Cell Stem Cell 8, 72–83 (2011)
Dou, D. R. et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat. Cell Biol. 18, 595–606 (2016)
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015)
Orkin, S. H. Controlling the fetal globin switch in man. Nature 301, 108–109 (1983)
Ciofani, M. & Zúñiga-Pflücker, J. C. The thymus as an inductive site for T lymphopoiesis. Annu. Rev. Cell Dev. Biol. 23, 463–493 (2007)
Alt, F. W. et al. VDJ recombination. Immunol. Today 13, 306–314 (1992)
Gillet, N. A. et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 117, 3113–3122 (2011)
North, T. E. et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 (2002)
Lacaud, G. et al. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood 100, 458–466 (2002)
Fernandes, I. et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11, 139–150 (2003)
Chan, F. C. et al. An RCOR1 loss-associated gene expression signature identifies a prognostically significant DLBCL subgroup. Blood 125, 959–966 (2015)
Nerlov, C. & Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 (1998)
Sauvageau, G., Iscove, N. N. & Humphries, R. K. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene 23, 7223–7232 (2004)
Ng, E. S. et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 34, 1168–1179 (2016)
Di-Poï, N., Koch, U., Radtke, F. & Duboule, D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Dev. Biol. 341, 488–498 (2010)
Lawrence, H. J. et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 106, 3988–3994 (2005)
Ng, A. P. et al. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood 115, 3966–3969 (2010)
Loughran, S. J. et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 9, 810–819 (2008)
Huang, Y. et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119, 388–398 (2012)
Weerkamp, F. et al. Identification of Notch target genes in uncommitted T-cell progenitors: no direct induction of a T-cell specific gene program. Leukemia 20, 1967–1977 (2006)
Hu, Z. et al. RUNX1 regulates corepressor interactions of PU.1. Blood 117, 6498–6508 (2011)
Sugimura, R. et al. Hematopoietic stem/progenitor cell conversion from human pluripotent stem cells http://dx.doi.org/10.1038/protex.2017.045 (2017)
Loh, Y. H. et al. Reprogramming of T cells from human peripheral blood. Cell Stem Cell 7, 15–19 (2010)
Park, I. H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008)
Meerbrey, K. L. et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl Acad. Sci. USA 108, 3665–3670 (2011)
Sankaran, V. G. et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature 460, 1093–1097 (2009)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nature Immunol. 14, 756–763 (2013)
Serrao, E., Cherepanov, P. & Engelman, A. N. Amplification, next-generation sequencing, and genomic DNA mapping of retroviral integration sites. J. Vis. Exp. (109): (2016)
Acknowledgements
We thank T. Schlaeger and the Boston Children’s Hospital Human ESC Core Facility, R. Mathieu from the flow cytometry core, R. Renee, A. Ratner, and S. Boswell from Harvard Medical School for RNA-seq, the orchestra team at Harvard Medical School for providing high-performance computing, D. Kaufman for providing the RUNX1c+24 hPSC line, N. Gerry for microarray and SNP array analysis, and T. North, C. Brendel and J. Powers for reading the manuscript. This work was supported by grants from the US National Institute of Diabetes and Digestive and Kidney Diseases (R24DK092760), the National Institute of Allergy and Infectious Diseases (R37AI039394), and the National Heart, Lung, Blood Institute Progenitor Cell Biology Consortium (UO1-HL100001) and the Progenitor Cell Translation Consortium (UO1-HL134812); Alex’s Lemonade Stand Foundation; and the Doris Duke Medical Foundation. G.Q.D. is an associate member of the Broad Institute and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. R.S. is supported by an American Society of Hematology Scholar Fellowship. C.S.V. is an EMBO (ALTF 1240-2015) fellow. S.D. is supported by the K99/R00HL123484. J.A.G. is supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK106311) and the Crohn’s and Colitis Foundation of America CDA 352644 (J.A.G.), S.B.S. is supported by NIDDK (DK034854), the Helmsley Charitable Trust, and the Wolpow Family Chair in IBD Treatment and Research.
Author information
Authors and Affiliations
Contributions
R.S. designed, performed, interpreted experiments, and wrote the paper. D.J., A.H., E.L.R., and T.N.Z. performed computational analysis of RNA-seq data. C.S.V., I.W., and P.S. assisted in iPSC culture, differentiation, and analyses of mice. C.S.V. replicated the entire process, independently. Y.L. performed globin expression analysis. R.R. performed cytospin of cells. M.M. performed flow cytometric analysis. D.J. contributed to writing and C.S.V. contributed to editing. E.S. and A.N.E. designed and interpreted integration sequencing experiments. J.A.G. and S.B.S. designed and interpreted TCRB rearrangement experiments. A.D. and G.K. instructed haemogenic endothelium induction. S.D. assisted in design and interpretation of experiments. G.Q.D. designed, interpreted experiments, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks B. Gottgens and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Induction of haemogenic endothelium from hPSCs.
a, Embryoid bodies formed from hPSCs. Subsequent panels show FACS analysis of day 8 embryoid bodies before magnetic cell isolation. b, CD34+FLK1+ population indicates haemogenic endothelium cells. CD235A and FLK1 plots show percentage of FLK1+CD235A− cells in embryoid bodies, further gated with CD43 and CD34 plots to detect haemogenic endothelium (CD34+FLK1+CD235A−CD43−). c, d, Haemogenic endothelium isolated at day 8 (Supplementary Fig. 1) was further cultured in EHT medium for the indicated number of days. qRT–PCR showed (c) downregulation of endothelial genes and (d) upregulation of haematopoietic genes. HSPC genes (RUNX1, SCL/TAL1) peak on day 3 of EHT culture; consequently, this time point was chosen for introducing transcription factors followed by transplantation in subsequent experiments. e, Microscopy and FACS analysis on day 7 of EHT showed the appearance of haematopoietic cells (CD34+CD45+). Data shown as mean ± s.d.
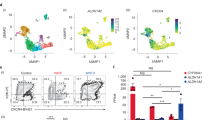
Extended Data Figure 2 Rationale for selecting candidate transcription factors for library screening.
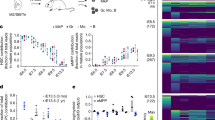
a, Heatmap of the expression profile of HSC-specific transcription factors (TFs) in haemogenic endothelium (CD34+FLK1+CD43−CD235A−) versus fetal-liver HSCs (CD34+CD38−CD90+CD45+). Twelve HSC-specific transcription factors enriched in fetal-liver HSCs relative to haemogenic endothelium (blue box) were cloned individually into a Dox-inducible lentiviral vector. b, The expression level of SOX17, a marker of haemogenic endothelium, was 2.4-fold higher in haemogenic endothelium (N = 7) than fetal-liver HSCs (N = 10). *P < 0.001. c, The library was supplemented with genes identified in previous screens. Candidates in red were drawn from a previous screen by the Rafii group3; blue from the Rossi group2; green from the Daley group7. The final library of 26 candidates is shown. d, Diagram of the Dox-ON pInducer-21 lentiviral vector used in this study (top). rtTA3 and eGFP are driven by EF1α-promoter; infection efficiency is indicated by the GFP signal. e, GFP analysis by FACS and fluorescence 3 days after infection of haemogenic endothelium cells. Routinely, over 50% transduction efficiency was achieved. For transplantation, haemogenic endothelium cells infected at day 3 EHT were incubated for 24 h and injected into mice. Dox was provided for 2 weeks in vivo after transplantation into sub-lethally irradiated immune-deficient NSG mice. f, Scheme for screening the 26 transcription factors library, and resulting haematopoietic chimaerism. hPSC-derived haemogenic endothelium was cultured for an additional 3 days in EHT medium, then infected with the library of 26 transcription factors. Infected cells (100,000) were injected intrafemorally into sub-lethally irradiated (250 rad) NSG mice, which were treated with doxycycline for 2 weeks to induce transgene expression in vivo. g, FACS analysis of bone marrow and thymus of an engrafted recipient is shown. Human CD45+ cells from bone marrow were analysed for CD33+ myeloid cells, CD19+ B cells, and CD3+ T cells as indicated. Thymic cells were analysed for human CD4 and CD8, with percentages of single and double-positive cells indicated. A photograph of an engrafted thymus is shown (bottom right). Data shown as mean ± s.d.
Extended Data Figure 3 Compiled data from transplantation experiments performed so far.
a, b, Total engrafted mice assessed in primary transplantation at various time points. a, Histogram of total mice injected with haemogenic endothelium cells and haemogenic endothelium cells infected with indicated library or constituent transcription factors. Haemogenic endothelium only: 2 out of 30 mice engrafted, none of which were multi-lineage (myeloid cells, erythroid cells, B cells and T cells); library: 11 out of 40 mice engrafted, 5 multi-lineage; 7 transcription factors: 33 out of 76 mice engrafted, 9 multi-lineage; 5TFpoly: 15 out of 30 mice engrafted, 5 multi-lineage. Engraftment was assessed from bone marrow at 4–16 weeks. b, Tables 1 and 2 show compiled data. c, Representative photomicrographs and FACS plots of robust EHT (top) and failed EHT (bottom). The appearance of round cells budding from adherent haemogenic endothelium cells by direct microscopic visualization was assessed routinely. RUNX1c+24 reporter positivity of haemogenic endothelium cells at day 3 of culture likewise reflected robustness of EHT (20% versus 5%).
Extended Data Figure 4 Identification of transcription factors that confer multi-lineage haematopoiesis in vivo.
a, SNP analysis of engrafted blood cells compared with hiPSC and cord blood MNCs. SNP array genotyping was conducted to confirm the origin of engrafted human cells from hiPSCs in representative mice. SNP genotypes for human CD45+ cells taken from bone marrow of engrafted mice, original hPSCs and reference cord blood MNCs were clustered, as shown, showing concordance of original hiPSCs and human cells recovered from bone marrow of engrafted mice. b, Tabular presentation of SNP data: concordance >99% indicates identity between cell types. Green highlight shows that original hPSC line (34hiPSC or H9 hESC) corresponds with human CD45+ cells in engrafted mice. Comparison with different hPSC lines or cord blood MNCs did not achieve 99% concordance, validating the SNP array as a means of defining origin of cells. c, Transgene detection in engrafted cells of primary recipients. CD33+ myeloid cells, CD19+ B cells, and CD3+ T cells were isolated from the human CD45+ population of bone marrow at 10 weeks from five independent mice. Genomic DNA of each cell type was analysed by PCR to detect integrated lentivirus. Identification number of recipients is shown (numbers 1, 5, and 6 were engrafted with hiPSC-derived haemogenic endothelium; numbers 2 and 3 were engrafted with hESC-derived haemogenic endothelium). L, left femur (injected side); R, right femur; +, positive control (lentiviral vector with each transcription factor). −, negative control (lentiviral vector without transcription factor). d, Overlap between transcription factors that conferred in vivo engraftment from hiPSC- and hESC-derived cells injected into mice and in vitro multi-lineage CFU potential. Transcription factors detected from genomic DNA PCR from in vitro colony screening and in vivo engraftment screening are shown. For in vivo screening, multiple cell lines (iPSC and ESC) were used. Factors detected by in vivo screening were overlapped with those detected by in vitro screening (RUNX1 and LCOR). Consistently, MYB and RORA were detected by in vitro screening as previously reported7. Overall, RUNX1, LCOR, SPI1, ERG, HOXA5, HOXA9, and HOXA10 (defined seven transcription factors) were identified in individual experiments with different PSC lines in vivo. e, Factor-minus-one approach to define essential transcription factors for engraftment. Haemogenic endothelium was infected with combination of seven transcription factors minus one each, as indicated, then transplanted into NSG mice. At the 8 week time point, engraftment of human CD45+ cells in bone marrow was determined by FACS. Each panel indicates a representative result of GFP vector: all seven transcription factors, and seven minus RUNX1, ERG, SPI1, LCOR, HOXA5, HOXA9, or HOXA10 as indicated in the panel. Reduction of chimaerism was seen when RUNX1, ERG, LCOR, HOXA5, or HOXA9 were removed. In contrast, omitting SPI1 or HOXA10 had a negligible effect on engraftment.
Extended Data Figure 5 FACS analysis after secondary transplantation of HE-7TF cells.
Human CD34+ cells were obtained by magnetic cell isolation from bone marrow of primary recipients of HE-7TF cells, then 3,000 cells (a, b) or 1,000 cells (c) were intrafemorally transplanted into secondary recipients. Multi-lineage engraftment in bone marrow from a representative recipient mouse is shown; a, 8 weeks; b, 14 weeks; c, 16 weeks. Specific mice analysed are indicated in Fig. 2b. d, Limiting dilution assay of HE-7TF cells after secondary transplantation. CD34+ cells were isolated from bone marrow of primary recipients, and either 1,000 or 3,000 cells were transplanted into secondary recipients. Multi-lineage engrafted recipients were counted as response. Confidence interval of 1/(stem-cell frequency) was calculated by ELDA (http://bioinf.wehi.edu.au/software/elda/) according to Poisson distribution. A limiting dilution assay of cord blood was used as reference. e, Transgene detection in engrafted cells of secondary recipients of HE-7TF cells. Recipient numbers are from Fig. 2b (numbers 31 and 35). f, Bone marrow chimaerism of primary mouse engrafted with 5-TFPoly at 12 weeks. Human CD45+ bone marrow of engrafted NSG was analysed for HSPCs (CD34+CD38−), nucleated erythroid (GLY-A+SYTO60+), enucleated erythroid (GLY-A+SYTO60−), neutrophils (PECAM+CD15+), B cells (IgM+CD19+), B progenitor cells (IgM−CD19+), B lymphocytes (IgM−CD19+CD38++), and T cells (CD3+/CD4, CD8). g, Representative FACS plots of bone marrow engrafted with human cord blood HSCs are shown at 10 and 12 weeks.
Extended Data Figure 6 Identification of the source of engraftable cells within the HE-7TF population.
a, Conversion of haemogenic endothelium (HE) into haematopoietic stem/progenitor cells by seven transcription factors requires EHT. Haemogenic endothelium or haemogenic endothelium grown in EHT medium for 3 days was transduced with seven transcription factors and transplanted into mice followed by bone marrow analysis at 4 weeks. Cells grown after 3 days of EHT showed CD45+ cells while those that were not grown under EHT conditions did not show CD45+ cells. b, Human umbilical vein endothelial cells were transduced with seven transcription factors or GFP lentiviral vectors, then cultured in EHT medium with Dox for a week. Flow cytometry analysis of PECAM (EC marker) and CD34 (haematopoietic marker) is shown. Human umbilical vein endothelial cells transduced with seven transcription factors fails to produce robust CD34+ cell population. c, Twenty-five thousand CD34+CD43+CD45+ (triple positive, TP) or CD34+CD43−CD45− (single positive, SP) cells were FACS-isolated and transplanted (N = 5 mice per group). Engraftment of human CD45+ cells was assessed in peripheral blood (PB) at 8 weeks. FACS plots of human CD45 and mouse CD45.1 of TP (left) and single positive (right) transplanted mice at the 8 week time point are shown. d, Multi-lineage engraftment of bone marrow and spleen from primary recipient mouse at 6 weeks is shown. CD34+CD43+CD45+ cells were intravenously injected.
Extended Data Figure 7 Molecular features of HE-7TF cells.
a, Correlation matrix of RNA-seq data from HSPC populations (CD34+CD38−CD45+) from HE-7TF cells or cord blood HSC engrafted for 12 weeks, iPS-haemogenic endothelium, and publicly available gene expression data (PubMed identifiers 26541609 and 26502406). RNA-seq samples from this study were RSips_7F_Average, RS_HE_Average, and RS_HSC_CB. Samples from ref. 23 were BCP_Average, CLP_Average, Thy_Average, HSC_Average, LMPP_Average, and HSC_CB_Average. The remaining samples are from ref. 24. b, Gene set enrichment analysis signature of HE-7TF cells compared with haemogenic endothelium cells. HE-7TF cells show gene expression signatures that positively correlate with LMO2 targets, TEF1 targets, HOXA4 targets, chemokine receptors and chemokines, TCF4 targets, integrin signal, GATA3 targets, and HSPCs. P < 0.05 and FDR q value < 0.25 were considered significant conditions. All gene set enrichment analysis plots satisfied these conditions, except the HSPC signatures, which had a q value of 0.121 and a P value of 0.06, suggesting that transcriptional differences remain between HE-7TF and bona fide HSCs/HSPCs. HSPC signature taken from ref. 56. c, RPKM values of seven transcription factors and HOXA target genes in indicated cell types are shown. HOXA target genes from ref. 28. PubMed identifier 27183470. d, Heatmap depiction of relative expression levels of the 26 transcription factors in the library in the following samples: HE, HE-7TF cells (engrafted), cord blood HSCs (engrafted), and fresh HSCs and progenitors. Notably, HE-7TF cells show high expression of HOXA family genes, GATA2, TGIF2, SOX4, and EVI1. e, The t-SNE of in-droplet single-cell RNA-seq of HE-7TF cells and cord blood HSCs (engrafted CD34+CD38−CD45+ cells from bone marrow at 12 weeks). The t-SNE from the top 500 most variable genes is presented in the top panel. f, Expression value of 8 haematopoietic genes in the same plot. Notably, the middle population (a subpopulation of HE-7TF cells) shared similar expression values, and degrees of heterogeneity of RUNX1, MEIS1, CD34, TAL1 with cord blood HSCs.
Extended Data Figure 8 Characterizations of differentiated cells: analysis of definitive erythropoiesis by relative quantification of globin transcripts.
a, Human GLY-A+ cells were isolated from lysed bone marrow (to exclude enucleated cells) and analysed by qRT–PCR to quantify (b) HBE, (c) HBG, and (d) HBB genes. CB, GLY-A+ erythroid cells from cord-blood-engrafted in NSG bone marrow; 7 TF HSPCs, GLY-A+ erythroid cells from seven transcription factor HSPC-engrafted in NSG bone marrow; 5F, GLY-A+ erythroid cells from hPSCs transduced with ERG, RORA, HOXA9, SOX4, and MYB7. Analysis of T-cell receptor diversity in engrafted T cells. e, Flow cytometric phenotyping of T cells from engrafted HE-7TF cells. Thymus was collected at 8 weeks and analysed for T-cell markers (CD4, CD8, CD3, TCRαβ, and TCRγδ). TCR phenotyping of the CD3+ population is shown on the right. One out of three recipients showed the presence of TCRγδ. Three thymic engrafted mice from independent experiments each. f–h, TCR rearrangement of thymocytes from cord blood CD34+ and HE-7TF engrafted in NSG. CD3+ T cells were isolated from NSG mice engrafted with (f) cord blood HSCs or (g) HE-7TF cells. Purified DNA was subjected to next-generation sequencing of the CDR3 using immunoSEQ (Adaptive Biotechnology) and analysed with the immunoSEQ Analyzer software (Adaptive Biotechnology). A high degree of combinatorial diversity in the V-gene segment usage was observed in CDR3 length, following a standard Gaussian distribution. h, Frequency of clonotype of T cells. i, Flow cytometric phenotyping of spleens from engrafted HE-7TF cells versus cord blood (j). Spleens were collected at 8 weeks and human CD45+ cells were analysed for T-cell markers (TCRβ, CD4, CD45RO, and CD45RA), and B-cell marker (CD19).
Extended Data Figure 9 Integration sequencing analysis of engrafted myeloid cells, B cells, and T cells from two individual animals.
a, Left: from 8 weeks, indicated in Fig. 2b; right: from 5 weeks. Clonally expanded populations are shown for each lineage, with common clones among three lineages represented by colour. The smallest proportional coloured segments for each animal represent the unit value of two clonal sequences. b, Overlap of commonly expanded integration sites from an animal shown on the left in a. Genomic DNA-sequencing of CD33+ myeloid cells, CD19+ B cells, and CD3+ T cells from bone marrow detected common expanded integration sites. c, Agarose gel electrophoresis of adaptor-ligation of engrafted cells. Cells were obtained independently from those used in a at the 1 week time point. d, Integrated loci mapped in genome. Nearby genes of common integrated sites are described. These data are from an animal indicated in a at the 8 week time point.
Extended Data Figure 10 Representative FACS plots.
a, b, FACS plot of cord blood HSCs (a) and HE-7TF cell-engrafted NSG mice at the 8 week time point (b). Each panel shows human CD45+ engraftment, HSPCs (CD34+CD38-), myeloid cells (CD33+), B cells (CD19+), T cells (CD3+CD4+CD8+), and erythrocytes (GLY-A+). Spleens were collected at 8 weeks, and human CD45+ cells were analysed for T-cell markers (TCRβ, CD4, CD45RO and CD45RA), and B-cell marker (CD19).
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, the uncropped gels and Supplementary Tables 1-2. (PDF 461 kb)
Rights and permissions
About this article
Cite this article
Sugimura, R., Jha, D., Han, A. et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017). https://doi.org/10.1038/nature22370
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22370
This article is cited by
-
Improving hematopoietic differentiation from human induced pluripotent stem cells by the modulation of Hippo signaling with a diarylheptanoid derivative
Stem Cell Research & Therapy (2024)
-
The adult environment promotes the transcriptional maturation of human iPSC-derived muscle grafts
npj Regenerative Medicine (2024)
-
Generating human bone marrow organoids for disease modeling and drug discovery
Nature Protocols (2024)
-
Nlrc3 signaling is indispensable for hematopoietic stem cell emergence via Notch signaling in vertebrates
Nature Communications (2024)
-
Modelling post-implantation human development to yolk sac blood emergence
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.