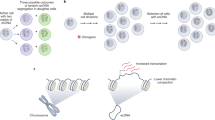
Abstract
Human cells have twenty-three pairs of chromosomes. In cancer, however, genes can be amplified in chromosomes or in circular extrachromosomal DNA (ecDNA), although the frequency and functional importance of ecDNA are not understood1,2,3,4. We performed whole-genome sequencing, structural modelling and cytogenetic analyses of 17 different cancer types, including analysis of the structure and function of chromosomes during metaphase of 2,572 dividing cells, and developed a software package called ECdetect to conduct unbiased, integrated ecDNA detection and analysis. Here we show that ecDNA was found in nearly half of human cancers; its frequency varied by tumour type, but it was almost never found in normal cells. Driver oncogenes were amplified most commonly in ecDNA, thereby increasing transcript level. Mathematical modelling predicted that ecDNA amplification would increase oncogene copy number and intratumoural heterogeneity more effectively than chromosomal amplification. We validated these predictions by quantitative analyses of cancer samples. The results presented here suggest that ecDNA contributes to accelerated evolution in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013)
Stark, G. R., Debatisse, M., Giulotto, E. & Wahl, G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57, 901–908 (1989)
Schimke, R. T. Gene amplification in cultured animal cells. Cell 37, 705–713 (1984)
Fan, Y. et al. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J. Appl. Genet. 52, 53–59 (2011)
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976)
McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015)
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012)
Yates, L. R. & Campbell, P. J. Evolution of the cancer genome. Nat. Rev. Genet. 13, 795–806 (2012)
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012)
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016)
Gillies, R. J., Verduzco, D. & Gatenby, R. A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 12, 487–493 (2012)
Von Hoff, D. D., Needham-VanDevanter, D. R., Yucel, J., Windle, B. E. & Wahl, G. M. Amplified human MYC oncogenes localized to replicating submicroscopic circular DNA molecules. Proc. Natl Acad. Sci. USA 85, 4804–4808 (1988)
Garsed, D. W. et al. The architecture and evolution of cancer neochromosomes. Cancer Cell 26, 653–667 (2014)
Carroll, S. M. et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol. Cell. Biol. 8, 1525–1533 (1988)
Windle, B., Draper, B. W., Yin, Y. X., O’Gorman, S. & Wahl, G. M. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 5, 160–174 (1991)
Kanda, T., Otter, M. & Wahl, G. M. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J. Cell Sci. 114, 49–58 (2001)
Mitelman, F., Johansson, B. & Mertens, F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman (2016)
Sanborn, J. Z. et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 73, 6036–6045 (2013)
Almendro, V. et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Reports 6, 514–527 (2014)
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013)
Nathanson, D. A. et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–76 (2014)
Storlazzi, C. T. et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 20, 1198–1206 (2010)
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010)
Li, X. et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett’s esophagus. Cancer Prev. Res. 7, 114–127 (2014)
Mishra, S. & Whetstine, J. R. Different facets of copy number changes: permanent, transient, and adaptive. Mol. Cell. Biol. 36, 1050–1063 (2016)
Schimke, R. T., Kaufman, R. J., Alt, F. W. & Kellems, R. F. Gene amplification and drug resistance in cultured murine cells. Science 202, 1051–1055 (1978)
Nikolaev, S. et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat. Commun. 5, 5690 (2014)
Biedler, J. L., Schrecker, A. W. & Hutchison, D. J. Selection of chromosomal variant in amethopterin-resistant sublines of leukemia L1210 with increased levels of dihydrofolate reductase. J. Natl. Cancer Inst. 31, 575–601 (1963)
Lee, P. M. Bayesian statistics: an introduction. 4th edn (John Wiley & Sons, 2012)
Motl, J. Bradley local image thresholding. https://www.mathworks.com/matlabcentral/fileexchange/40854 (2015)
Bradley, D. & Roth, G. Adaptive thresholding using the integral image. J. Graphics Tools 12, 13–21 (2007)
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Kent, W. J. et al. The human genome browser at UCSC. Genome Res 12, 996–1006 (2002)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Miller, C. A., Hampton, O., Coarfa, C. & Milosavljevic, A. ReadDepth: a parallel R package for detecting copy number alterations from short sequencing reads. PLoS One 6, e16327 (2011)
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016)
Acknowledgements
We thank R. Kolodner, W. Mischel, D. Geschwind, members of the Mischel laboratory, A. Akbari, A. Iranmehr and A. Patel for helpful comments. This work was supported by the Ludwig Institute for Cancer Research (P.S.M., B.R., K.A., W.K.C., F.B.F.), Defeat GBM Program of the National Brain Tumor Society (P.S.M., F.B.F.), The Ben and Catherine Ivy Foundation (P.S.M.), generous donations from the Ziering Family Foundation in memory of Sigi Ziering (P.S.M.); The Susan G. Komen Foundation (SAC110036), The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002) and The Breast Cancer Research Foundation (BCRF) to G.M.W.; CureSearch for Children’s Cancer and a Leadership Award from the California Institute for Regenerative Medicine to R.W.R. This work was also supported by the following NIH grants: NS73831 (P.S.M.), GM114362 (V.B., V.D., D.B.), NS80939 (F.B.F.), CA014195 and CA159859 (G.M.W.) and CA151819 (D.A.N.) and T32CA121938 (K.M.T.) and NSF grants: NSF-IIS-1318386 and NSF-DBI-1458557 (V.B., V.D., D.B.).
Author information
Authors and Affiliations
Contributions
K.M.T., V.D., D.B., V.B. and P.S.M. conceived and designed the study. K.M.T., V.D., D.B., T.K., J.R., C.L. and B.L. performed experiments. D.B., V.D. and V.B. developed the ECdetect software and performed mathematical modelling and simulations. V.D. and V.B. developed the AmpliconArchitect software. K.A., B.R., D.A.N., F.B.F., W.K.C., P.N.R. and G.M.W. provided analytic support. H.I.K., M.D.T., S.K, R.W.-R. and S.R.V. provided additional clinical samples and analytic support. K.M.T., V.D., D.B., V.B. and P.S.M. wrote the manuscript with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
V.B. is a co-founder, has an equity interest and receives income from Digital Proteomics, LLC. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. Digital Proteomics was not involved in the research presented here.
Additional information
Reviewer Information: Nature thanks C. Maley, A. Papenfuss and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 2 Alternative analysis of ecDNA presence according to varying criteria, stratified by sample type.
Samples with a minimum number of ecDNA elements per 10 cells in metaphase in average shown in x axis are classified ecDNA positive, and their fraction is displayed on the y axis. The vertical line at x = 4 shows that for a minimum of 4 ecDNA elements per 10 cells in metaphase on average, 0% of normal, 10% of immortalized, 46% of tumour cell line and 89% of PDX samples are classified as ecDNA positive.
Extended Data Figure 4 Histogram of depth of coverage for next-generation sequencing of tumour samples.
We sequenced 117 tumour samples including 63 cell lines, 19 neurospheres (PDX) and 35 cancer tissues with coverage ranging from 0.6× to 3.89× (excluding one sample with 0.06× coverage) with median coverage of 1.19×.
Extended Data Figure 5 Full metaphase spreads corresponding to the partial metaphase spreads shown in Fig. 3c.
Extended Data Figure 7 Copy-number amplification and diversity due to ecDNA.
To test how much of the copy-number amplification and diversity could be attributed to ecDNA, we chose FISH probes that bind to four of the most commonly amplified oncogenes in our sample set, EGFR, MYC, CCND1 or ERBB2, and quantified the cell-to-cell variability in their DNA copy number in metaphase spreads, from four tumour cell lines: GBM39, MB411FH, SF295 and PC3 cancer cells. For each cell line, only the target oncogene marked in red is known to be amplified on ecDNA (EGFR in GBM39; MYC in MB411FH and PC3, and CCND1 in SF295). The other 3 genes reside on chromosomal loci. The target oncogene shows consistently higher copy numbers (top) and diversity (bottom).
Extended Data Figure 8 Fine structure analysis of EGFRvIII amplification in extrachromosomal or chromosomal DNA in GBM39 cells.
a, FISH images showed the EGFR gene on ecDNA (top) and HSRs (bottom) in different passages of the GBM39 cell line. Analysis of the HSR FISH images shows evidence of multiple integration sites on different chromosomes. b, Next-generation sequencing of DNA from 4 independent cultures of GBM39 was used to analyse the fine structure of amplifications (Supplementary Information 4.3). In 3 biological replicates (rows 1–3) of these cultures, EGFRvIII was exclusively on ecDNA, whereas one of the later passage cultures (row 4) was found to contain EGFRvIII entirely on HSRs, with no detectable ecDNA. The DNA derived from different ecDNA cultures shows identical structure with some heterogeneity (P < 2.18 × 10−8 for all pairs), suggesting common origin. However, DNA derived from HSRs show a conserved structure that is identical to the ecDNA structure (P < 1.98 × 10−5, Supplementary Information 2.4), possibly with tandem duplications. c, A possible progression of normal genome to cancer genome with EGFRvIII ecDNA and amplification to a copy count of around 100 copies. The EGFRvIII ecDNA elements possibly aggregate into tandem duplications and reintegrate into multiple chromosomes as HSRs, so that 5–6 HSRs accommodate around 100 copies of EGFRvIII.
Extended Data Figure 9 Fine structure analysis of EGFRvIII amplification in extrachromosomal or chromosomal DNA in naive GBM39 cells and in response to erlotinib treatment and drug withdrawal.
a, FISH images of naive GBM39 cells, in response to erlotinib treatment (ERZ) and drug withdrawal (after ERZ is removed) displayed ecDNA amplification, HSR amplification and ecDNA amplification, respectively (top to bottom). b, Next-generation sequencing of DNA from 6 independent cultures of GBM39 was used to analyse the fine structure of amplifications (Supplementary Information 4.3). Average copy numbers of amplified intervals as determined from sequencing analysis in naive samples: 110–150 (biological replicates in rows 1–3), the ERZ sample: 5.4 (row 4) and after drug withdrawal: 100–105 (biological replicates in rows 5–6). All three categories show similar fine structure indicating common origin (Methods). Drug-withdrawal replicates show additional rearrangements and heterogeneity as compared to naive samples. c, Cytogenetic and sequencing progression suggests that the EGFRvIII ecDNA in naive cells get reintegrated into HSRs after drug application and that the copies in the HSRs break off from the chromosomes again to form ecDNA with a copy count similar to naive cells. Drug-withdrawal samples also show additional heterogeneity in structure.
Supplementary information
Supplementary Information
This file contains Supplementary sections 1-4 comprising: Section 1 ECDNA count and presence statistics; Section 2 ECdetect - Software for detection of extrachromosomal DNA from DAPI staining metaphase images; Section 3 AmpliconArchitect: Sequence analysis for identification and reconstruction of focal amplifications; Section 4 A theoretical model of extrachromosomal and intrachromosomal duplication and Supplementary References. (PDF 14810 kb)
Rights and permissions
About this article
Cite this article
Turner, K., Deshpande, V., Beyter, D. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017). https://doi.org/10.1038/nature21356
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21356
This article is cited by
-
Mechanistic insights and the clinical prospects of targeted therapies for glioblastoma: a comprehensive review
Experimental Hematology & Oncology (2024)
-
ABCB1 overexpression through locus amplification represents an actionable target to combat paclitaxel resistance in pancreatic cancer cells
Journal of Experimental & Clinical Cancer Research (2024)
-
Evolving copy number gains promote tumor expansion and bolster mutational diversification
Nature Communications (2024)
-
scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells
Nature Communications (2024)
-
Extrachromosomal DNA in cancer
Nature Reviews Cancer (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.