Abstract
B-lymphoid transcription factors, such as PAX5 and IKZF1, are critical for early B-cell development1,2, yet lesions of the genes encoding these transcription factors occur in over 80% of cases of pre-B-cell acute lymphoblastic leukaemia (ALL)3,4. The importance of these lesions in ALL has, until now, remained unclear. Here, by combining studies using chromatin immunoprecipitation with sequencing and RNA sequencing, we identify a novel B-lymphoid program for transcriptional repression of glucose and energy supply. Our metabolic analyses revealed that PAX5 and IKZF1 enforce a state of chronic energy deprivation, resulting in constitutive activation of the energy-stress sensor AMPK5,6,7. Dominant-negative mutants of PAX5 and IKZF1, however, relieved this glucose and energy restriction. In a transgenic pre-B ALL mouse model, the heterozygous deletion of Pax5 increased glucose uptake and ATP levels by more than 25-fold. Reconstitution of PAX5 and IKZF1 in samples from patients with pre-B ALL restored a non-permissive state and induced energy crisis and cell death. A CRISPR/Cas9-based screen of PAX5 and IKZF1 transcriptional targets identified the products of NR3C1 (encoding the glucocorticoid receptor)8, TXNIP (encoding a glucose-feedback sensor)9 and CNR2 (encoding a cannabinoid receptor)10 as central effectors of B-lymphoid restriction of glucose and energy supply. Notably, transport-independent lipophilic methyl-conjugates of pyruvate and tricarboxylic acid cycle metabolites bypassed the gatekeeper function of PAX5 and IKZF1 and readily enabled leukaemic transformation. Conversely, pharmacological TXNIP and CNR2 agonists and a small-molecule AMPK inhibitor strongly synergized with glucocorticoids, identifying TXNIP, CNR2 and AMPK as potential therapeutic targets. Furthermore, our results provide a mechanistic explanation for the empirical finding that glucocorticoids are effective in the treatment of B-lymphoid but not myeloid malignancies. Thus, B-lymphoid transcription factors function as metabolic gatekeepers by limiting the amount of cellular ATP to levels that are insufficient for malignant transformation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
30 May 2018
In the NR3C1 plot of Fig. 3c (left) of this Letter, the curves for TXNIP (middle) were inadvertently plotted. This has been corrected online, and see Supplementary Information to the Amendment for the original Fig. 3c.
References
Nutt, S. L., Heavey, B., Rolink, A. G. & Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 (1999)
Georgopoulos, K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994)
Mullighan, C. G. et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 (2007)
Mullighan, C. G. et al. BCR–ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453, 110–114 (2008)
Nakada, D., Saunders, T. L. & Morrison, S. J. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658 (2010)
Gan, B. et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704 (2010)
Gurumurthy, S. et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659–663 (2010)
Brennan-Speranza, T. C. et al. Osteoblasts mediate the adverse effects of glucocorticoids on fuel metabolism. J. Clin. Invest. 122, 4172–4189 (2012)
Wu, N. et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell 49, 1167–1175 (2013)
Nogueiras, R. et al. The endocannabinoid system: role in glucose and energy metabolism. Pharmacol. Res. 60, 93–98 (2009)
Medina, K. L. et al. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell 7, 607–617 (2004)
Di Tullio, A. et al. CCAAT/enhancer binding protein alpha (C/EBPα)-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl Acad. Sci. USA 108, 17016–17021 (2011)
Fretz, J. A. et al. Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology 151, 1611–1621 (2010)
Wu, Z. et al. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3, 151–158 (1999)
Shaw, R. J. et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl Acad. Sci. USA 101, 3329–3335 (2004)
Martín-Lorenzo, A. et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discov. 5, 1328–1343 (2015)
Faubert, B. et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 17, 113–124 (2013)
Zhou, G. et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 (2001)
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014)
Pui, C. H. & Evans, W. E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 354, 166–178 (2006)
Hanus, L. et al. HU-308: a specific agonist for CB2, a peripheral cannabinoid receptor. Proc. Natl Acad. Sci. USA 96, 14228–14233 (1999)
Stoltzman, C. A., Kaadige, M. R., Peterson, C. W. & Ayer, D. E. MondoA senses non-glucose sugars: regulation of thioredoxin-interacting protein (TXNIP) and the hexose transport curb. J. Biol. Chem. 286, 38027–38034 (2011)
Foley, S. B. et al. Expression of BCR/ABL p210 from a knockin allele enhances bone marrow engraftment without inducing neoplasia. Cell Rep. 5, 51–60 (2013)
Papaemmanuil, E. et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6–RUNX1 acute lymphoblastic leukemia. Nat. Genet. 46, 116–125 (2014)
Swaminathan, S. et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat. Immunol. 16, 766–774 (2015)
Cazzaniga, G. et al. Developmental origins and impact of BCR–ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood 118, 5559–5564 (2011)
Wiemels, J. L. et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 354, 1499–1503 (1999)
Bose, S., Deininger, M., Gora-Tybor, J., Goldman, J. M. & Melo, J. V. The presence of typical and atypical BCR–ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood 92, 3362–3367 (1998)
Damm, F. et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 4, 1088–1101 (2014)
Li, S., Ilaria, R. L., Jr, Million, R. P., Daley, G. Q. & Van Etten, R. A. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189, 1399–1412 (1999)
Xie, H., Ye, M., Feng, R. & Graf, T. Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 (2004)
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015)
Ochiai, K. et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 13, 300–307 (2012)
Thai, M. et al. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 19, 694–701 (2014)
Harvey, R. C. et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116, 4874–4884 (2010)
Kang, H. et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood 115, 1394–1405 (2010)
Liu, G. J. et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev. 28, 1337–1350 (2014)
Holmfeldt, L. et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45, 242–252 (2013)
Acknowledgements
This work was supported by the NIH/NCI through Outstanding Investigator Award R35CA197628 (to M.M.), R01CA137060, R01CA157644 and R01CA172558 (to M.M.), a Wellcome Trust Senior Investigator Award and a Leukemia and Lymphoma Scholar award (to M.M.), the Howard Hughes Medical Institute HHMI-55108547 (to M.M.), the Alex’s Lemonade Stand Foundation for Childhood Cancer (to M.M.), the William Lawrence & Blanche Hughes Foundation for childhood cancer (to M.M.), the Norman and Sadie Lee Foundation (for Pediatric Cancer, to M.M.), the Falk Trust through a Falk Medical Research Trust Catalyst Award (to M.M.), Cancer Research Institute (CRI) through a Clinic and Laboratory Integration Program (CLIP) grant (to M.M.), the Melanoma Research Alliance Established Investigator Award (to T.G.G.), the German Bundesministerium für Bildung und Forschung, BMBF (to A.H.) and the German Carreras Foundation (DJCLS R13/26) (to A.B. and I.S.-G.). T.G.G. is an American Cancer Society Research Scholar. M.M. is a Howard Hughes Medical Institute (HHMI) Faculty Scholar.
Author information
Authors and Affiliations
Contributions
M.M. conceived the study. L.N.C. and M.M. wrote the paper and designed experiments. L.N.C., Z.C., D.B., J.-W.L., G.X., K.N.C., C.H., S.S. and V.C. performed experiments and analysed data. S.M.K. performed functional proteomics. H.G. performed biostatistical analyses. T.A.M., T.E., A.H., M.K. and G.C. provided and characterized patient samples. I.S.-G., A.B., T.S.R., G.J.L. and R.A.D. provided mouse models. M.A.P., K.R.Y., G.J.L., R.A.D., I.S.-G., A.B., H.P.K. and H.S. provided specific expertise in NR3C1, PAX5 and IKZF1 function. D.B. and T.G.G. performed experimentation and provided analysis and expertise in metabolomics.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
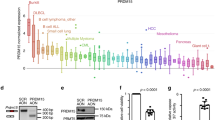
Extended Data Figure 1 Frequent genetic lesions of B-lymphoid transcription factors in B-cell lineage leukaemia.
a, Gene expression of B-lymphoid transcription factors (top) as well as positive (middle) and negative (bottom) regulators of glucose uptake and energy supply upon inducible restoration of Pax5 (GEO accession number: GSE52870)38 in haploinsufficient pre-B leukaemia cells. b, Lesions in PAX5, IKZF1, EBF1 and TCF3 were studied in clinical trials for B-lymphoid ALL in children (P9906; n = 187; top) and adults (MDACC; n = 92; bottom). Red and grey boxes denote patient samples with detected lesions. c, Protein expression of PAX5 and IKZF1 was examined by western blot in ten patient-derived pre-B ALL samples. The control samples were CD19+ B cells from four healthy donors. d, ChIP–seq analysis for binding of B-lymphoid transcription factors in human B cells (ENCODE cell ID: GM12878) to promoter regions of molecules implicated in positive (INSR, GLUT1, GLUT3, GLUT6, HK2, G6PD, LKB1) and negative (NR3C1, TXNIP, CNR2) regulation of glucose uptake and utilization. e, Recruitment of PAX5 was confirmed by quantitative ChIP in patient-derived pre-B ALL cells. Data shown as mean ± s.d. from three independent experiments and assessed by two-tailed t-test. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2 The B-lymphoid transcription factor PAX5 functions as a metabolic gatekeeper.
a, Protein levels of PAX5 and IKZF1 in haploinsufficient patient-derived pre-B ALL cells and pre-B ALL cells expressing functional PAX5 and IKZF1. b, Number of viable cells and cell viability upon inducible activation of PAX5 in haploinsufficient patient-derived pre-B ALL (PAX5∆) cells. c, To test whether Pax5 functions as metabolic gatekeeper, BCR–ABL1-induced changes in glycolytic activity and capacity (ECAR), glucose uptake, and ATP levels (normalized to cell numbers) were studied in Pax5 wild-type and Pax5 haploinsufficient pre-B cells in the presence or absence of a BCR–ABL1-transgene. Data shown as mean ± s.d. from three independent experiments and assessed by two-tailed t-test (b, left; c) or two-way ANOVA (b, right). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 Divergent metabolic characteristics of myeloid and B-lineage leukaemia.
a, Glycolytic reserve (ECAR) and mitochondrial functions (OCR) in patient-derived myeloid (CML) and B-lymphoid (Ph+ ALL) leukaemia samples (n = 5, each in triplicate). Values were normalized to total protein and are shown as mean ± s.d., assessed by two-tailed t-test. b, Mouse pre-B ALL cells were reprogrammed into myeloid differentiation using a doxycycline-inducible TetOn-Cebpa vector system, and characterized by flow cytometry (representative results from three independent experiments). c, Heat map of gene expression of glucose uptake and metabolism regulators (GEO accession number: GSE32330)12. d, Western blots of mouse pre-B ALL cells upon B-to-myeloid cell reprogramming to verify gene expression changes. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4 The energy stress sensor LKB1–AMPK plays a pro-survival role and modulates glucose uptake and energy supply in pre-B ALL.
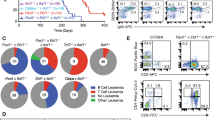
a, Lkb1fl/fl mice were crossed with Cre-deleter strains for deletion at early pre-B-cell stages (Mb1) and in fully mature B cells (Cd21). B-cell populations in bone marrow and spleen (n = 3 litter mates) were characterized by flow cytometry analysis. b, The catalytic subunit of Ampk has two isoforms, α1 and α2. Analysis of published gene expression data (GSE38463)38 revealed that expression of the α1-form peaks at later stages of B-cell development, whereas expression of both Lkb1 and the α2-form of Ampk peaks in pre-B cells. For this reason, we studied the consequences of inducible ablation of Lkb1 and Ampka2 in mouse models of BCR–ABL1-transformed pre-B ALL cells. Protein levels of Lkb1 and Ampkα2 were verified by western blots. c, Viable cell counts upon Cre-mediated deletion of Lkb1 or Ampka2 or on treatment with an empty vector (EV). d, Apoptosis following Lkb1 deletion was monitored by annexin V/ 7-aminoactinomycin D (7AAD) staining. e, Colony-forming ability was assessed by serial re-plating upon deletion of Lkb1 in pre-B ALL cells. f, Glucose uptake and ATP levels (normalized to cell numbers) were measured following Cre-mediated deletion of Ampka2. g, Luciferase bioimaging of transplant recipient mice injected with Lkb1fl/fl pre-B ALL cells transduced with 4-OHT-inducible Cre or an empty vector and treated with tamoxifen (0.4 mg per mouse; n = 7 per group). h, Overall survival was assessed by a Kaplan–Meier analysis (P value calculated by Mantel–Cox log-rank test). i, Leukaemia samples developed in recipient mice (Fig. 2d) were genotyped for the presence of either floxed or deleted Lkb1 and Ampka2 alleles (n = 3 mice). Representative FACS plots and images from three independent experiments are shown (a, d, e). Data shown as mean ± s.d. from three independent experiments and assessed by two-way ANOVA (c) or two-tailed t-test (e, f). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5 LKB1 and AMPK are independent predictors of poor clinical outcome for patients with pre-B ALL.
a, b, Children with high-risk pre-B ALL (P9906, n = 207) were divided into two groups based on higher- or lower-than-median mRNA levels of LKB1 (a) or AMPKΑ2 (b). c, d, Adults with acute myeloid leukaemia (AML; The Cancer Genome Atlas, n = 184) were divided into two groups based on higher- or lower-than-median mRNA levels of LKB1 (c) or AMPKΑ2 (d). Overall survival of patients was assessed in the two groups by Kaplan–Meier analysis. A log-rank test was used to assess statistical significance (a–d). e, Frequencies of somatic mutations in the coding regions of EBF1 (top, blue), IKZF1 (middle, grey) and PAX5 (bottom, red) from the Catalogue of Somatic Mutations in Cancer (COSMIC) database are plotted for pre-B ALL and various subtypes of mature B-cell lymphoma. Somatic mutations in the 5′ UTR regions of EBF1, IKZF1 and PAX5 frequently represent by-products of somatic hypermutation during normal B-cell development and are not included in this analysis. PCNL, primary central nervous system lymphoma.
Extended Data Figure 6 Divergent functions of Lkb1 in BCR–ABL1-driven pre-B ALL and myeloid leukaemia.
a, Staining of Lkb1fl/fl BCR–ABL1 myeloid (CML-like) and B-lineage (Ph+ ALL-like) leukaemia cells with (Cre) or without (empty vector (EV)) deletion of Lkb1 for the surface markers CD19, B220 (B-lymphoid), Sca-1 and CD13 (myeloid). b, Phosphorylation of AMPKα-T172, AKT-S473 and S6-S235 and protein levels of cell-cycle checkpoint molecules Arf, p53 and p27 following Lkb1-deletion. c, Viable cell counts upon deletion of Lkb1. d, Glucose uptake and ATP levels (normalized to cell numbers) in myeloid and B-lymphoid leukaemia cells upon Lkb1-deletion (n = 6). Data are shown as mean ± s.d. e, Cell-cycle analyses were performed by measuring BrdU incorporation in combination with 7AAD staining. Percentages of cells in G0/1, S, and G2/M phases are shown. Viability following Lkb1 deletion was monitored by annexin V/7AAD staining. Representative FACS plots from three independent experiments are shown (a, e). Data shown as mean ± s.d. from three independent experiments (c, e), assessed by two-tailed t-test (c–e). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7 Activity of the energy-stress sensor LKB1 represents a specific vulnerability of B-lymphoid leukaemia.
a, b, Glycolytic profiles (ECAR; a) and mitochondrial functions (OCR; b) upon Lkb1-deletion in B-lymphoid leukaemia cells with (left) or without (right) B-to-myeloid cell reprogramming. Values were normalized to total protein (n = 6). c, AMP levels in sorted B-lymphoid and B-to-myeloid reprogrammed cells are shown as log2-transformed relative amounts (the amount in Lkb1-deleted cells divided by the average amount in control samples), and data were baseline-centred (the baseline is equal to the average amount in control samples; n = 3). d, Phosphorylation of Ampkα-T172, Ulk-S555, Raptor-S792 and Acc-S79 was assessed by western blots. Data shown as mean ± s.d. and assessed by two-tailed t-test (a–c). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 8 Small molecule inhibition of AMPK in human pre-B ALL.
a, Human leukaemia and lymphoma cells (n = 4 biological replicates for B-cell lymphoma and pre-B ALL; n = 3 biological replicates for CML; each in triplicate) were treated with BML275 (72 h), and relative viability was assessed. b, Apoptosis was examined by annexin V/7AAD staining in patient-derived pre-B ALL samples (n = 3) upon treatment with BML275 (10 μmol/l). c, Phospho-ACC-S79 in patient-derived pre-B ALL samples (n = 5) following overnight treatment with control (−) or BML275 (+, 10 μmol/l) was assessed. d, Phosphorylation of S6-S235/236 and Akt-S473 in patient-derived pre-B ALL samples (n = 6) following overnight treatment with control (−) or BML275 (+, 10 μmol/l) was assessed. e, Levels of AMP (normalized to cell numbers) in patient-derived CML (CML5 and CML6) and Ph+ ALL (ICN1 and PDX2) cells following treatment with control or BML275 (10 μmol/l for 12 h). Plotted are log2-transformed average relative amounts (amount in cells treated with BML275 divided by the average amount of that in control cells; two cases per group; each in triplicate); data were median-centred. Median-centring was performed separately for CML5, CML6, ICN1 and PDX2 samples. f, Patient-derived CML (left) and pre-B ALL (middle) cells as well as MYC-driven B-cell lymphoma cell lines (right) were treated with BML275 (10 μmol/l) or vehicle control for 6 h. Glycolytic profiles (ECAR) and mitochondrial functions (OCR) were measured (normalized to total protein; n = 6; f). Data shown as mean ± s.d. and assessed by two-tailed t-test (e, f).
Extended Data Figure 9 Mechanistic contribution of PAX5 targets.
a, b, Representative FACS plots from CRISPR-based gene editing experiments (a). CRISPR complexes were delivered to patient-derived PAX5-haploinsufficient pre-B ALL cells along with RFP-tagged gRNAs to direct dCas9 (CRISPR-mediated gene activation) or Cas9 (CRISPR-mediated gene deletion) to specific PAX5 target genes (for example, NR3C1; left). Upon inducible activation of GFP-tagged PAX5 or an empty vector (EV) in patient-derived pre-B ALL cells, enrichment or depletion of GFP+ cells carrying RFP-tagged gRNAs (RFP+) was monitored by flow cytometry (right; NT, non-targeting; g-53, gRNA clone 53 for deletion of NR3C1). Changes in percentage of GFP+ cells carrying the indicated gRNAs following induction, as compared to cells carrying the non-targeting gRNA (b). c, d, In mouse pre-B ALL models for genetic loss of Nr3c1, Cnr2, and Txnip function, responses to prednisolone (Pred) were measured (c) and protein levels of Nr3c1, Txnip and Cnr2 were examined (d). e, In a patient-derived PAX5-haploinsufficient pre-B ALL sample (PAX5Δ; left), dexamethasone responses upon inducible activation of PAX5 or empty vector were measured. In a patient-derived PAX5 wild-type pre-B ALL sample (PAX5WT; right), effects of DN-PAX5 on dexamethasone responses were measured. Likewise, dose–response curves for dexamethasone were measured in two patient-derived pre-B ALL samples carrying either wild-type or deleted IKZF1 upon induction of DN-IKZF1 or IKZF1 expression, respectively. Data shown as mean ± s.d. from three independent experiments and assessed by two-tailed t-test (b) or two-way ANOVA (c, e).
Extended Data Figure 10 Targeting AMPK, CNR2 and TXNIP in combination with glucocorticoids.
a, Three different patient-derived pre-B ALL samples were treated with the AMPK inhibitor BML275, prednisolone or a combination of the two for 72 h. b, Three different patient-derived pre-B ALL samples were treated with the CNR2 agonist HU308, prednisolone (Pred), or a combination of the two for 72 h. c, d, Three different patient-derived pre-B ALL samples were treated with the TXNIP agonist 3-OMG (c) or d-allose (d), prednisolone, or a combination of the two for 72 h. Relative viability was assessed (a–d). Combination index (CI) values at ED50 are shown. Prednisolone concentrations used were twofold higher than those of BML275. All data shown as mean ± s.d. (n = 3 independent experiments).
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, which shows the gel scans for the Figures and Extended Data Figures related to this paper, Supplementary Tables 1- 10 and additional references. (PDF 2530 kb)
Source data
Rights and permissions
About this article
Cite this article
Chan, L., Chen, Z., Braas, D. et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 542, 479–483 (2017). https://doi.org/10.1038/nature21076
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21076
This article is cited by
-
Cannabinoids induce cell death in leukaemic cells through Parthanatos and PARP-related metabolic disruptions
British Journal of Cancer (2024)
-
Should I stay or should I go? Spatio-temporal control of cellular anchorage by hematopoietic factors orchestrates tumor metastatic cascade
Molecular Cancer (2023)
-
Disruption to the FOXO-PRDM1 axis resulting from deletions of chromosome 6 in acute lymphoblastic leukaemia
Leukemia (2023)
-
The role of TXNIP in cancer: a fine balance between redox, metabolic, and immunological tumor control
British Journal of Cancer (2023)
-
Pediatric T-cell acute lymphoblastic leukemia blast signature and MRD associated immune environment changes defined by single cell transcriptomics analysis
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.