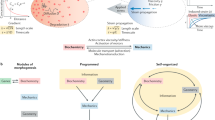
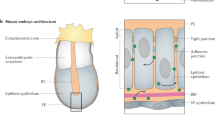
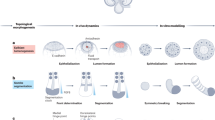
Abstract
A long-term aim of the life sciences is to understand how organismal shape is encoded by the genome. An important challenge is to identify mechanistic links between the genes that control cell-fate decisions and the cellular machines that generate shape, therefore closing the gap between genotype and phenotype. The logic and mechanisms that integrate these different levels of shape control are beginning to be described, and recently discovered mechanisms of cross-talk and feedback are beginning to explain the remarkable robustness of organ assembly. The 'full-circle' understanding of morphogenesis that is emerging, besides solving a key puzzle in biology, provides a mechanistic framework for future approaches to tissue engineering.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Thomasen, A. L. “Historia animalium” compared to “Gynaecia” in the literature of the Middle Ages. Clio Med. 15, 5–24 (1980).
Keller, R. Physical biology returns to morphogenesis. Science 338, 201–203 (2012).
Heisenberg, C.-P. & Bellaïche, Y. Forces in tissue morphogenesis and patterning. Cell 153, 948–962 (2013).
Nüsslein-Volhard, C. & Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). The first systematic study on the genetic regulation of embryonic development; it remains unparalleled in impact and importance.
Lawrence, P. A. Morphogens: how big is the big picture? Nature Cell Biol. 3, E151–E154 (2001).
Green, J. B. A. & Sharpe, J. Positional information and reaction-diffusion: two big ideas in developmental biology combine. Development 142, 1203–1211 (2015).
Munro, E. M. & Odell, G. M. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development 129, 13–24 (2002).
Dawes-Hoang, R. E. et al. Folded gastrulation, cell shape change and the control of myosin localization. Development 132, 4165–4178 (2005).
Lecuit, T. Adhesion remodeling underlying tissue morphogenesis. Trends Cell Biol. 15, 34–42 (2005).
Settleman, J. & Baum, B. Cell shape and tissue morphogenesis. Semin. Cell Dev. Biol. 19, 213–214 (2008).
Zallen, J. A. & Blankenship, J. T. Multicellular dynamics during epithelial elongation. Semin. Cell Dev. Biol. 19, 263–270 (2008).
Barrallo-Gimeno, A. & Nieto, M. A. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132, 3151–3161 (2005).
Revenu, C. & Gilmour, D. EMT 2.0: shaping epithelia through collective migration. Curr. Opin. Genet. Dev. 19, 338–342 (2009).
Thiery, J.-P., Acloque, H., Huang, R. Y. J. & Nieto, M. A. Epithelial–mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Friedl, P., Locker, J., Sahai, E. & Segall, J. E. Classifying collective cancer cell invasion. Nature Cell Biol. 14, 777–783 (2012).
Friedl, P. & Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nature Rev. Mol. Cell Biol. 10, 445–457 (2009).
Keller, R. E. An experimental analysis of the role of bottle cells and the deep marginal zone in gastrulation of Xenopus laevis. J. Exp. Zool. 216, 81–101 (1981).
Martin, A. C. & Goldstein, B. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141, 1987–1998 (2014).
Leptin, M. & Grunewald, B. Cell shape changes during gastrulation in Drosophila. Development 110, 73–84 (1990).
Sweeton, D., Parks, S., Costa, M. & Wieschaus, E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development 112, 775–789 (1991).
Martin, A. C., Kaschube, M. & Wieschaus, E. F. Pulsed contractions of an actin–myosin network drive apical constriction. Nature 457, 495–499 (2009). This paper shows that apical constriction in the Drosophila mesoderm is driven by a medially localized pulsatile actin–myosin network in incremental, contractile steps.
Mason, F. M., Tworoger, M. & Martin, A. C. Apical domain polarization localizes actin–myosin activity to drive ratchet-like apical constriction. Nature Cell Biol. 15, 926–936 (2013).
Vasquez, C. G., Tworoger, M. & Martin, A. C. Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J. Cell Biol. 206, 435–450 (2014).
Solon, J., Kaya-Çopur, A., Colombelli, J. & Brunner, D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell 137, 1331–1342 (2009).
Kölsch, V., Seher, T., Fernandez-Ballester, G. J., Serrano, L. & Leptin, M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science 315, 384–386 (2007). This study identifies a target of Twist called T48 that recruits RhoGEF2 specifically to the apical membrane in ventral-furrow cells and elucidated the interplay between Rho1 signalling and Snail in the apical relocalization of adherens junctions.
Weng, M. & Wieschaus, E. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. J. Cell Biol. 212, 219–229 (2016).
Sawyer, J. K., Harris, N. J., Slep, K. C., Gaul, U. & Peifer, M. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186, 57–73 (2009).
Oda, H., Tsukita, S. & Takeichi, M. Dynamic behavior of the cadherin-based cell–cell adhesion system during Drosophila gastrulation. Dev. Biol. 203, 435–450 (1998).
Leptin, M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 5, 1568–1576 (1991).
Nieto, M. A. Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850 (2013).
Chanet, S. & Schweisguth, F. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nature Cell Biol. 14, 467–476 (2012).
Mathew, S. J., Rembold, M. & Leptin, M. Role for Traf4 in polarizing adherens junctions as a prerequisite for efficient cell shape changes. Mol. Cell. Biol. 31, 4978–4993 (2011).
Costa, M., Wilson, E. T. & Wieschaus, E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell 76, 1075–1089 (1994).
Manning, A. J., Peters, K. A., Peifer, M. & Rogers, S. L. Regulation of epithelial morphogenesis by the G protein-coupled receptor mist and its ligand fog. Sci. Signal. 6, ra98 (2013). Refs 34 and 63 report the identification of the long sought-after receptors for Fog, which function additively in the mesoderm.
Kim, H. Y., Varner, V. D. & Nelson, C. M. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development 140, 3146–3155 (2013).
Plageman, T. F. et al. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development 137, 405–415 (2010).
Plageman, T. F. et al. A Trio–RhoA–Shroom3 pathway is required for apical constriction and epithelial invagination. Development 138, 5177–5188 (2011).
Eiraku, M., Adachi, T. & Sasai, Y. Relaxation–expansion model for self-driven retinal morphogenesis. Bioessays 34, 17–25 (2012).
Haigo, S. L., Hildebrand, J. D., Harland, R. M. & Wallingford, J. B. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13, 2125–2137 (2003).
Hildebrand, J. D. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118, 5191–5203 (2005).
Nishimura, T. & Takeichi, M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 135, 1493–1502 (2008).
Hildebrand, J. D. & Soriano, P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99, 485–497 (1999).
Lee, C., Le, M. P. & Wallingford, J. B. The shroom family proteins play broad roles in the morphogenesis of thickened epithelial sheets. Dev. Dyn. 238, 1480–1491 (2009).
Lang, R. A., Herman, K., Reynolds, A. B., Hildebrand, J. D. & Plageman, T. F. p120-catenin-dependent junctional recruitment of Shroom3 is required for apical constriction during lens pit morphogenesis. Development 141, 3177–3187 (2014).
Chung, M.-I., Nascone-Yoder, N. M., Grover, S. A., Drysdale, T. A. & Wallingford, J. B. Direct activation of Shroom3 transcription by Pitx proteins drives epithelial morphogenesis in the developing gut. Development 137, 1339–1349 (2010).
Ernst, S. et al. Shroom3 is required downstream of FGF signalling to mediate proneuromast assembly in zebrafish. Development 139, 4571–4581 (2012).
Das, D. et al. The interaction between Shroom3 and Rho-kinase is required for neural tube morphogenesis in mice. Biol. Open 3, 850–860 (2014).
Irvine, K. D. & Wieschaus, E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841 (1994).
Bertet, C., Sulak, L. & Lecuit, T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671 (2004). Using the Drosophila embryo, this paper reveals a mechanism of cell intercalation that was subsequently shown to drive tissue elongation in a variety of organs and organisms.
Blankenship, J. T., Backovic, S. T., Sanny, J. S. P., Weitz, O. & Zallen, J. A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459–470 (2006).
Lienkamp, S. S. et al. Vertebrate kidney tubules elongate using a planar cell polarity–dependent, rosette-based mechanism of convergent extension. Nature Genet. 44, 1382–1387 (2012).
Williams, M., Yen, W., Lu, X. & Sutherland, A. Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev. Cell 29, 34–46 (2014).
Rozbicki, E. et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nature Cell Biol. 17, 397–408 (2015).
Shih, J. & Keller, R. Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development 116, 901–914 (1992).
Keller, R. et al. Mechanisms of convergence and extension by cell intercalation. Phil. Trans. R. Soc. Lond. B 355, 897–922 (2000).
Tada, M. & Heisenberg, C.-P. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development 139, 3897–3904 (2012).
Shindo, A. & Wallingford, J. B. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652 (2014).
Zallen, J. A. & Wieschaus, E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355 (2004).
Rauzi, M., Verant, P., Lecuit, T. & Lenne, P.-F. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nature Cell Biol. 10, 1401–1410 (2008).
Fernandez-Gonzalez, R. et al. Dynamics are regulated by tension in intercalating cells. Dev. Cell 17, 736–743 (2009).
Paré, A. C. et al. A positional Toll receptor code directs convergent extension in Drosophila. Nature 515, 523–527 (2014). The patterned expression of three Toll receptor family members is shown to link anterior–posterior tissue patterning to cellular behaviour that drives cell intercalation and germband elongation.
Nishimura, T., Honda, H. & Takeichi, M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149, 1084–1097 (2012).
Kerridge, S. et al. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nature Cell Biol. 18, 261–270 (2016).
Wang, Y.-C., Khan, Z., Kaschube, M. & Wieschaus, E. F. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature 484, 390–393 (2012). This paper shows that epithelial folding in the Drosophila embryo can be initiated by the basal movement of adherens junctions, independently of myosin activity.
Turner, F. R. & Mahowald, A. P. Scanning electron microscopy of Drosophila melanogaster embryogenesis: II. gastrulation and segmentation. Dev. Biol. 57, 403–416 (1977).
Vincent, A., Blankenship, J. T. & Wieschaus, E. Integration of the head and trunk segmentation systems controls cephalic furrow formation in Drosophila. Development 124, 3747–3754 (1997).
Spencer, A. K., Siddiqui, B. A. & Thomas, J. H. Cell shape change and invagination of the cephalic furrow involves reorganization of F-actin. Dev. Biol. 402, 192–207 (2015).
Barrett, K., Leptin, M. & Settleman, J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell 91, 905–915 (1997).
Aigouy, B. et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773–786 (2010).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Collinet, C., Rauzi, M., Lenne, P.-F. & Lecuit, T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nature Cell Biol. 17, 1247–1258 (2015).
Rauzi, M. et al. Embryo-scale tissue mechanics during Drosophila gastrulation movements. Nature Commun. 6, 8677 (2015).
Desprat, N., Supatto, W., Pouille, P., Beaurepaire, E. & Farge, E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev. Cell 15, 470–477 (2008).
Butler, L. C. et al. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nature Cell Biol. 11, 859–864 (2009).
Lye, C. M. et al. Mechanical coupling between endoderm invagination and axis extension in Drosophila. PLoS Biol. 13, e1002292 (2015).
Martin, A. C., Gelbart, M., Fernandez-Gonzalez, R., Kaschube, M. & Wieschaus, E. F. Integration of contractile forces during tissue invagination. J. Cell Biol. 188, 735–749 (2010).
Spahn, P. & Reuter, R. A vertex model of Drosophila ventral furrow formation. PLoS ONE 8, e75051 (2013).
Hashimoto, H., Robin, F. B., Sherrard, K. M. & Munro, E. M. Sequential contraction and exchange of apical junctions drives zippering and neural tube closure in a simple chordate. Dev. Cell 32, 241–255 (2015).
Alexandre, C., Baena-Lopez, A. & Vincent, J.-P. Patterning and growth control by membrane-tethered Wingless. Nature 505, 180–185 (2014).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Kornberg, T. B. Cytonemes and the dispersion of morphogens. Wiley Interdiscip. Rev. Dev. Biol. 3, 445–463 (2014).
Averbukh, I., Ben-Zvi, D., Mishra, S. & Barkai, N. Scaling morphogen gradients during tissue growth by a cell division rule. Development 141, 2150–2156 (2014).
Snijder, B. & Pelkmans, L. Origins of regulated cell-to-cell variability. Nature Rev. Mol. Cell Biol. 12, 119–125 (2011).
Curto, M., Cole, B. K., Lallemand, D., Liu, C.-H. & McClatchey, A. I. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 177, 893–903 (2007). Refs 84–90 provide mechanistic insights into how changes in cell contact and density can feed back into the activity of key signalling pathways.
Frechin, M. et al. Cell-intrinsic adaptation of lipid composition to local crowding drives social behaviour. Nature 523, 88–91 (2015).
Etoc, F. et al. A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev Cell 39, 302–315 (2016).
Klingner, C. et al. Isotropic actomyosin dynamics promote organization of the apical cell cortex in epithelial cells. J. Cell Biol. 207, 107–121 (2014).
Haag, A. et al. An in vivo EGF receptor localization screen in C. elegans identifies the Ezrin homolog ERM-1 as a temporal regulator of signaling. PLoS Genet. 10, e1004341 (2014).
Nallet-Staub, F. et al. Cell density sensing alters TGF-β signaling in a cell-type-specific manner, independent from Hippo pathway activation. Dev. Cell 32, 640–651 (2015).
Narimatsu, M., Samavarchi-Tehrani, P., Varelas, X. & Wrana, J. L. Distinct polarity cues direct Taz/Yap and TGFβ receptor localization to differentially control TGFβ-induced Smad signaling. Dev. Cell 32, 652–656 (2015).
Wang, H. et al. Rap–GEF signaling controls stem cell anchoring to their niche through regulating DE-Cadherin-mediated cell adhesion in the Drosophila testis. Dev. Cell 10, 117–126 (2006).
Michel, M., Raabe, I., Kupinski, A. P., Pérez-Palencia, R. & Bökel, C. Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nature Commun. 2, 415 (2011).
Matusek, T. et al. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature 516, 99–103 (2014).
Harmansa, S., Hamaratoglu, F., Affolter, M. & Caussinus, E. Dpp spreading is required for medial but not for lateral wing disc growth. Nature 527, 317–322 (2015).
Nechiporuk, A. & Raible, D. W. FGF-dependent mechanosensory organ patterning in zebrafish. Science 320, 1774–1777 (2008).
Lecaudey, V., Cakan-Akdogan, G., Norton, W. H. J. & Gilmour, D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development 135, 2695–2705 (2008).
Harding, M. J. & Nechiporuk, A. V. Fgfr–Ras–MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development 139, 3130–3135 (2012).
Durdu, S. et al. Luminal signalling links cell communication to tissue architecture during organogenesis. Nature 515, 120–124 (2014). Refs 98 and 99 reveal crucial feedback roles for 3D tissue architecture in regulating cell fate and behaviour during organogenesis in vivo.
Shyer, A. E., Huycke, T. R., Lee, C., Mahadevan, L. & Tabin, C. J. Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569–580 (2015).
Karlsson, L., Lindahl, P., Heath, J. K. & Betsholtz, C. Abnormal gastrointestinal development in PDGF-A and PDGFR-α deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457–3466 (2000).
Discher, D. E., Janmey, P. & Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005).
Bellas, E. & Chen, C. S. Forms, forces, and stem cell fate. Curr. Opin. Cell Biol. 31, 92–97 (2014).
Chanet, S. & Martin, A. C. Mechanical force sensing in tissues. Prog. Mol. Biol. Transl. Sci. 126, 317–352 (2014).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). Refs 104 and 105 elegantly demonstrate that changes in substrate mechanics can influence lineage decisions in cultured stem cells.
McBeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K. & Chen, C. S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 (2004).
Swift, J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013).
Buckley, C. D. et al. The minimal cadherin–catenin complex binds to actin filaments under force. Science 346, 1254211 (2014).
Mouilleron, S., Langer, C. A., Guettler, S., McDonald, N. Q. & Treisman, R. Structure of a pentavalent G-actin•MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci. Signal. 4, ra40 (2011).
Sero, J. E. et al. Cell shape and the microenvironment regulate nuclear translocation of NF-κB in breast epithelial and tumor cells. Mol. Syst. Biol. 11, 790 (2015).
Farge, E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr. Biol. 13, 1365–1377 (2003).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). The transcriptional regulators YAP and TAZ are shown to act as sensors and mediators of mechanical inputs, therefore providing a mechanism by which force can control gene expression and fate.
Wei, S. C. et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nature Cell Biol. 17, 678–688 (2015).
Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 (2014).
Maître, J.-L. et al. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 536, 344–348 (2016). This study uncovers a role for mechanics in regulating the first cell-fate decision in the early mouse embryo.
Brunet, T. et al. Evolutionary conservation of early mesoderm specification by mechanotransduction in Bilateria. Nature Commun. 4, 2821 (2013).
Benham-Pyle, B. W., Pruitt, B. L. & Nelson, W. J. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015).
Saha, A. et al. Determining physical properties of the cell cortex. Biophys. J. 110, 1421–1429 (2016).
Etournay, R. et al. TissueMiner: a multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. eLife 5, e14334 (2016).
Bielmeier, C. et al. Interface contractility between differently fated cells drives cell elimination and cyst formation. Curr. Biol. 26, 563–574 (2016).
Morelli, L. G., Uriu, K., Ares, S. & Oates, A. C. Computational approaches to developmental patterning. Science 336, 187–191 (2012).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). A groundbreaking study that describes the self-assembly of embryonic stem cells into optic-cup organoids in the absence of pre-patterned chemical or mechanical gradients.
Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Davies, J. A. & Cachat, E. Synthetic biology meets tissue engineering. Biochem. Soc. Trans. 44, 696–701 (2016).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791 (2016).
Eiraku, M., Adachi, T. & Sasai, Y. Relaxation–expansion model for self-driven retinal morphogenesis. Bioessays 34, 17–25 (2012).
Turner, D. A., Baillie-Johnson, P. & Martinez Arias, A. Organoids and the genetically encoded self-assembly of embryonic stem cells. Bioessays 38, 181–191 (2016).
Parks, S. & Wieschaus, E. The Drosophila gastrulation gene concertina encodes a Gα-like protein. Cell 64, 447–458 (1991).
Kanesaki, T., Hirose, S., Grosshans, J. & Fuse, N. Heterotrimeric G protein signaling governs the cortical stability during apical constriction in Drosophila gastrulation. Mech. Dev. 130, 132–142 (2013).
Homem, C. C. F. & Peifer, M. Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development 135, 1005–1018 (2008).
Fox, D. T. & Peifer, M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development 134, 567–578 (2006).
Hampoelz, B., Hoeller, O., Bowman, S. K., Dunican, D. & Knoblich, J. A. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nature Cell Biol. 7, 1099–1105 (2005).
Fuse, N., Yu, F. & Hirose, S. Gprk2 adjusts Fog signaling to organize cell movements in Drosophila gastrulation. Development 140, 4246–4255 (2013).
Spahn, P., Ott, A. & Reuter, R. The PDZ-GEF protein Dizzy regulates the establishment of adherens junctions required for ventral furrow formation in Drosophila. J. Cell Sci. 125, 3801–3812 (2012).
Sawyer, J. K. et al. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell 22, 2491–2508 (2011).
Zhang, Y. et al. The glucosyltransferase Xiantuan of the endoplasmic reticulum specifically affects E-Cadherin expression and is required for gastrulation movements in Drosophila. Dev. Biol. 390, 208–220 (2014).
Wang, Y.-C., Khan, Z. & Wieschaus, E. F. Distinct Rap1 activity states control the extent of epithelial invagination via β-catenin. Dev. Cell 25, 299–309 (2013).
Menzies, A. S. Mena and vasodilator-stimulated phosphoprotein are required for multiple actin-dependent processes that shape the vertebrate nervous system. J. Neurosci. 24, 8029–8038 (2004).
Nakajima, H. & Tanoue, T. Epithelial cell shape is regulated by Lulu proteins via myosin-II. J. Cell Sci. 123, 555 (2010).
Simões, S. de M. et al. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell 19, 377–388 (2010).
Munjal, A., Philippe, J.-M., Munro, E. & Lecuit, T. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature 524, 351–355 (2015).
Tamada, M., Farrell, D. L. & Zallen, J. A. Abl regulates planar polarized junctional dynamics through β-catenin tyrosine phosphorylation. Dev. Cell 22, 309–319 (2012).
Levayer, R., Pelissier-Monier, A. & Lecuit, T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nature Cell Biol. 13, 529–540 (2011).
Simões, S. D. M., Mainieri, A. & Zallen, J. A. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 204, 575–589 (2014).
Turing, A. M. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 (1952).
Acknowledgements
We are grateful to S. De Renzis, F. Peri and the Gilmour and Leptin groups for discussions. We thank C. Böckel, S. Durdu, A. Shyer and T. Hiiragi for help with Fig. 3.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Reviewer Information Nature thanks C.-P. Heisenberg and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Rights and permissions
About this article
Cite this article
Gilmour, D., Rembold, M. & Leptin, M. From morphogen to morphogenesis and back. Nature 541, 311–320 (2017). https://doi.org/10.1038/nature21348
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21348
This article is cited by
-
How multiscale curvature couples forces to cellular functions
Nature Reviews Physics (2024)
-
Proliferation-driven mechanical compression induces signalling centre formation during mammalian organ development
Nature Cell Biology (2024)
-
Dynamic interplay of microtubule and actomyosin forces drive tissue extension
Nature Communications (2024)
-
Synthetic morphology with agential materials
Nature Reviews Bioengineering (2023)
-
Homeotic compartment curvature and tension control spatiotemporal folding dynamics
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.