Abstract
Although lysine acetylation is now recognized as a general protein modification for both histones and non-histone proteins1,2,3, the mechanisms of acetylation-mediated actions are not completely understood. Acetylation of the C-terminal domain (CTD) of p53 (also known as TP53) was an early example of non-histone protein acetylation4 and its precise role remains unclear. Lysine acetylation often creates binding sites for bromodomain-containing ‘reader’ proteins5,6. Here we use a proteomic screen to identify the oncoprotein SET as a major cellular factor whose binding with p53 is dependent on CTD acetylation status. SET profoundly inhibits p53 transcriptional activity in unstressed cells, but SET-mediated repression is abolished by stress-induced acetylation of p53 CTD. Moreover, loss of the interaction with SET activates p53, resulting in tumour regression in mouse xenograft models. Notably, the acidic domain of SET acts as a ‘reader’ for the unacetylated CTD of p53 and this mechanism of acetylation-dependent regulation is widespread in nature. For example, acetylation of p53 also modulates its interactions with similar acidic domains found in other p53 regulators including VPRBP (also known as DCAF1), DAXX and PELP1 (refs. 7, 8, 9), and computational analysis of the proteome has identified numerous proteins with the potential to serve as acidic domain readers and lysine-rich ligands. Unlike bromodomain readers, which preferentially bind the acetylated forms of their cognate ligands, the acidic domain readers specifically recognize the unacetylated forms of their ligands. Finally, the acetylation-dependent regulation of p53 was further validated in vivo by using a knock-in mouse model expressing an acetylation-mimicking form of p53. These results reveal that acidic-domain-containing factors act as a class of acetylation-dependent regulators by targeting p53 and, potentially, other proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 (2010)
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009)
Kim, S. C. et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 (2006)
Gu, W. & Roeder, R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997)
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999)
Marmorstein, R. & Zhou, M. M. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 (2014)
Kim, K. et al. Vpr-binding protein antagonizes p53-mediated transcription via direct interaction with H3 tail. Mol. Cell. Biol. 32, 783–796 (2012)
Zhao, L. Y. et al. Negative regulation of p53 functions by Daxx and the involvement of MDM2. J. Biol. Chem. 279, 50566–50579 (2004)
Nair, B. C. et al. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ. 21, 1409–1418 (2014)
Li, T. et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012)
Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015)
Simeonova, I. et al. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Reports 3, 2046–2058 (2013)
Hamard, P. J. et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 27, 1868–1885 (2013)
von Lindern, M. et al. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 12, 3346–3355 (1992)
Kim, J. Y. et al. Inhibition of p53 acetylation by INHAT subunit SET/TAF-Iβ represses p53 activity. Nucleic Acids Res. 40, 75–87 (2012)
Tang, Z. et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 154, 297–310 (2013)
Jin, Q. et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011)
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009)
Vousden, K. H. & Prives, C. Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009)
Berger, S. L. Keeping p53 in check: a high-stakes balancing act. Cell 142, 17–19 (2010)
Matsumoto, K., Nagata, K., Okuwaki, M. & Tsujimoto, M. Histone- and chromatin-binding activity of template activating factor-I. FEBS Lett. 463, 285–288 (1999)
Kim, K. B. et al. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Iβ regulates Ku70-mediated DNA damage response. Cell. Mol. Life Sci. 71, 2731–2745 (2014)
Chae, Y. C. et al. Inhibition of FoxO1 acetylation by INHAT subunit SET/TAF-Iβ induces p21 transcription. FEBS Lett. 588, 2867–2873 (2014)
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Cohen, H. Y. et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell 13, 627–638 (2004)
Daitoku, H. et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl Acad. Sci. USA 101, 10042–10047 (2004)
Krummel, K. A., Lee, C. J., Toledo, F. & Wahl, G. M. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc. Natl Acad. Sci. USA 102, 10188–10193 (2005)
Feng, L., Lin, T., Uranishi, H., Gu, W. & Xu, Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 25, 5389–5395 (2005)
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015)
Li, Y. et al. Accurate in silico identification of species-specific acetylation sites by integrating protein sequence-derived and functional features. Sci. Rep. 4, 5765 (2014)
Acknowledgements
We thank F. Giancotti, X. Yang, R. K. Vadlamudi and W. An for providing reagents for this work. We also thank R. Baer for discussion and suggestions. This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5R01CA193890, 5RO1CA190477, 5RO1CA085533 and 2P01CA080058 to W.G. and GM030518 and CA121852 to B.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by D.W., N.K., G.L. and W.G. The experiments were performed mainly by D.W. and N.K. Bioinformatic analysis was performed by G.L. Mass spectrometry analysis was performed by W.L. The xenograft assay was performed by D.W. and L.J. Data were analysed and interpreted by D.W., N.K., G.L., W.-G.Z., J.Q., B.H. and W.G. The manuscript was written by D.W., N.K., G.L. and W.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
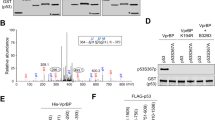
Extended Data Figure 1 Further analysis of p53–SET interaction.
a, A list of SET peptides identified by mass spectrometry. b, In vitro binding assay of methylated p53 CTD and purified SET. c–e, In vitro binding assay between SET and the purified ubiquitinated, sumoylated or neddylated forms of p53. f, g, Western blot analysis of p53 and SET domains for their interaction. In vitro binding assay was performed by incubating immobilized GST, GST–p53 or GST–SET with each purified SET or p53 protein, as indicated. h, Western blot analysis of the interaction between p53 and SET in cells. H1299 cells were co-transfected with indicated constructs and the nuclear extract was analysed by co-IP assay. i–k, ChIP analysis of p53 or SET recruitment onto the PUMA (i), TIGAR (j) or GLS2 (k) promoter. HCT116 cells were treated with or without 1 μM doxorubicin for 24 h and then the cellular extracts were analysed by ChIP assay with indicated antibodies. Asterisks indicate the specific bands of indicated proteins. Error bars indicate mean ± s.d., n = 3 for technical replicates. Data are shown as representative of three experiments. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 2 RNA-seq analysis to identify genes regulated by p53–SET interplay.
a, Western blot analysis of the expression of p53 in U2OS-derived CRISPR control cells or CRISPR p53-KO cells. b, Heat map of genes regulated by the p53–SET interplay. U2OS (CRISPR Ctr or CRISPR p53-KO) cells were transfected with control siRNA or SET-specific siRNA for 4 days and the total RNA was prepared for RNA-seq analysis with two or three biological replicates, as indicated. Known p53 target genes which were also repressed by SET in a p53-dependent manner were selected and presented as a heat map. The relative SET expression is shown in the last row of the heat map. c, qPCR validation of the genes regulated by the p53–SET interplay. Error bars indicate mean ± s.d., n = 3 for technical replicates. Data are shown as representative of three experiments. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 3 SET-mediated effects on cell proliferation and tumour growth.
a, b, Representative image (a) or quantitative analysis (b) of the SET knockdown-mediated effect on cell growth of U2OS-derived CRISPR control cells or CRISPR p53-KO cells. c, Western blot analysis of the expression of p53 in HCT116-derived CRISPR control cells or CRISPR p53-KO cells. d, Xenograft analysis of the SET-mediated effect on tumour growth by HCT116-derived CRISPR control cells or CRISPR p53-KO cells. e, Western blot analysis of p53 expression in control or derived HCT116 cell lines, as indicated. Error bars indicate mean ± s.d., n = 3 in b or n = 5 in d for biological replicates. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 4 SET regulates histone modifications on p53 target promoter.
a, Western blot analysis of the SET knockdown-mediated effect on the p53 C-terminal acetylation in HCT116 cells. Doxorubicin (Dox)-treated cells were also analysed in parallel as a positive control. b, Western blot analysis of the SET-mediated effect on the CBP-induced p53 C-terminal acetylation in H1299 cells. c, e, ChIP analysis of promoter-recruitment of p53 (c) or p300/CBP (e) upon SET depletion in HCT116 cells. d, ChIP analysis of the SET-knockdown-mediated effect on histone modifications in the PUMA promoter in HCT116 cells. f, ChIP analysis of the SET-mediated effect on p53-dependent H3K18 and H3K27 acetylation in the PUMA promoter. Error bars indicate mean ± s.d., n = 3 for technical replicates. Data are shown as representative of three experiments. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 5 Acetylation regulates the interaction between acidic-domain-containing proteins and their acetylatable ligands.
a, A summary table of characteristic features of the acidic-domain-containing proteins SET, VPRBP, DAXX and PELP1. The acidic amino acids are underlined. b, In vitro binding assay of p53 CTD and purified full-length VPRBP, DAXX or PELP1. c–e, Western blot analysis of the interaction between p53 and VPRBP (c), DAXX (d) or PELP1 (e) in the nuclear fraction of H1299 cells. f–h, In vitro binding assay between purified SET and KRD of H3 (f), KU70 (g) or FOXO1 (h). i, In vitro binding assay of the H3 KRD and purified VPRBP, DAXX or PELP1. j, In vitro binding assay of the H3 KRD and BRD4 or BRD7 (nuclear extract). Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 6 p53KQ mutant mimics acetylated p53.
a, Schematic diagram of human unacetylated p53 and the acetylation-deficient and acetylation-mimicking mutants of p53. b, In vitro binding assay of SET and different types of p53, as indicated. c–e, Western blot analysis of the interaction between acidic-domain-containing proteins (c, VPRBP; d, DAXX; e, PELP1) and different types of p53 in cells. H1299 cells were co-transfected with indicated constructs, and the nuclear extract was analysed by Co-IP assay. Asterisks indicate the purified proteins. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 7 Generation of p53KQ/KQ mice.
a, Schematic diagram of the gene targeting strategy to replace the p53 C-terminal 7 lysines with 7 glutamines in mouse p53. b, Southern blot screening of ES cells to identify p53+/KQ clones. c, PCR genotyping analysis of wild-type (110 bp), p53+/KQ heterozygous (110 bp and 150 bp), and p53KQ/KQ homozygous mice (150 bp only). d, Sequencing analysis of the transcripts prepared from the p53+/KQ heterozygous mouse spleen. e, A summary table of observed numbers of mice from p53+/KQ heterozygous intercrosses. f, Positive control for p53 staining in the IHC assay. The spleen tissue sections of p53+/+ mice treated with or without 6 Gy γ-radiation was stained with p53 (CM-5) antibody. g, h, Representative image (g) or quantitative analysis (h) of SET-knockdown-mediated cell growth of p53+/+ or p53KQ/KQ MEFs (P2). Error bars indicate mean ± s.d., n = 3 for biological replicates. Uncropped blots can be found in Supplementary Fig. 1.
Extended Data Figure 8 Characterization of Set conditional knockout mice.
a, Schematic diagram of the strategy to generate Set conditional knockout mice. b, Validation of Set knockout in embryos (E8.5) by genotyping and western blot analysis. c, A summary table of observed numbers of embryos or pups from Set+/− intercrosses. d, Representative pictures of Set+/+ and Set−/− embryos (E10.5). e, qPCR analysis of the expression of p53 target genes in Set+/+ and Set−/− embryos (E10.5). Error bars indicate mean ± s.d., n = 3 for technical replicates. Data are shown as representative of three experiments. Uncropped blots can be found in Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-2 and Supplementary Tables. (PDF 1475 kb)
Rights and permissions
About this article
Cite this article
Wang, D., Kon, N., Lasso, G. et al. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538, 118–122 (2016). https://doi.org/10.1038/nature19759
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19759
This article is cited by
-
Oncoprotein SET-associated transcription factor ZBTB11 triggers lung cancer metastasis
Nature Communications (2024)
-
The CDK4/6 Inhibitor Palbociclib Induces Cell Senescence of High-grade Serous Ovarian Cancer Through Acetylation of p53
Biochemical Genetics (2024)
-
KLF12 promotes the proliferation of breast cancer cells by reducing the transcription of p21 in a p53-dependent and p53-independent manner
Cell Death & Disease (2023)
-
Specific regulation of BACH1 by the hotspot mutant p53R175H reveals a distinct gain-of-function mechanism
Nature Cancer (2023)
-
α-Catenin acetylation is essential for its stability and blocks its tumor suppressor effects in breast cancer through Yap1
Cancer Gene Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.