Abstract
Mitochondrial electron transport chain complexes are organized into supercomplexes responsible for carrying out cellular respiration. Here we present three architectures of mammalian (ovine) supercomplexes determined by cryo-electron microscopy. We identify two distinct arrangements of supercomplex CICIII2CIV (the respirasome)—a major ‘tight’ form and a minor ‘loose’ form (resolved at the resolution of 5.8 Å and 6.7 Å, respectively), which may represent different stages in supercomplex assembly or disassembly. We have also determined an architecture of supercomplex CICIII2 at 7.8 Å resolution. All observed density can be attributed to the known 80 subunits of the individual complexes, including 132 transmembrane helices. The individual complexes form tight interactions that vary between the architectures, with complex IV subunit COX7a switching contact from complex III to complex I. The arrangement of active sites within the supercomplex may help control reactive oxygen species production. To our knowledge, these are the first complete architectures of the dominant, physiologically relevant state of the electron transport chain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-8130, EMD-8128 and EMD-8129. The models have been deposited in the Protein Data Bank (PDB) under accession codes 5J4Z, 5J7Y and 5J8K.
References
Sazanov, L. A. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nature Rev. Mol. Cell Biol. 16, 375–388 (2015)
Chance, B., Estabrook, R. W. & Lee, C. P. Electron transport in the oxysome. Science 140, 379–380 (1963)
Hatefi, Y., Haavik, A. G. & Griffiths, D. E. Studies on the electron transfer system. XL. Preparation and properties of mitochondrial DPNH-coenzyme Q reductase. J. Biol. Chem. 237, 1676–1680 (1962)
Chazotte, B. & Hackenbrock, C. R. The multicollisional, obstructed, long-range diffusional nature of mitochondrial electron transport. J. Biol. Chem. 263, 14359–14367 (1988)
Hackenbrock, C. R., Chazotte, B. & Gupte, S. S. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J. Bioenerg. Biomembr. 18, 331–368 (1986)
Schägger, H. & Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000)
Schägger, H. & Pfeiffer, K. The ratio of oxidative phosphorylation complexes I–V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 276, 37861–37867 (2001)
Acín-Pérez, R. et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 (2004)
Maranzana, E., Barbero, G., Falasca, A. I., Lenaz, G. & Genova, M. L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 19, 1469–1480 (2013)
Bianchi, C., Genova, M. L., Parenti Castelli, G. & Lenaz, G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J. Biol. Chem. 279, 36562–36569 (2004)
Blaza, J. N., Serreli, R., Jones, A. J. Y., Mohammed, K. & Hirst, J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc. Natl Acad. Sci. USA 111, 15735–15740 (2014)
Enríquez, J. A. Supramolecular organization of respiratory complexes. Annu. Rev. Physiol. 78, 533–561 (2016)
Lenaz, G., Tioli, G., Falasca, A. I. & Genova, M. L. Complex I function in mitochondrial supercomplexes. BBA Bioenergetics 1857, 991–1000 (2016)
Lapuente-Brun, E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013)
Ikeda, K., Shiba, S., Horie-Inoue, K., Shimokata, K. & Inoue, S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nature Commun. 4, 2147 (2013)
Mourier, A., Matic, S., Ruzzenente, B., Larsson, N.-G. & Milenkovic, D. The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab. 20, 1069–1075 (2014)
Williams, E. G. et al. Systems proteomics of liver mitochondria function. Science 352, aad0189 (2016)
Althoff, T., Mills, D. J., Popot, J. L. & Kühlbrandt, W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1 . EMBO J. 30, 4652–4664 (2011)
Dudkina, N. V., Kudryashev, M., Stahlberg, H. & Boekema, E. J. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl Acad. Sci. USA 108, 15196–15200 (2011)
Mileykovskaya, E. et al. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J. Biol. Chem. 287, 23095–23103 (2012)
Mileykovskaya, E. & Dowhan, W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids 179, 42–48 (2014)
Xia, D. et al. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. BBA Bioenergetics 1827, 1278–1294 (2013)
Vinothkumar, K. R., Zhu, J. & Hirst, J. Architecture of mammalian respiratory complex I. Nature 515, 80–84 (2014)
Tsukihara, T. et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272, 1136–1144 (1996)
Pitceathly, R. D. S. et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Reports 3, 1795–1805 (2013)
Balsa, E. et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 16, 378–386 (2012)
Hayashi, T. et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc. Natl Acad. Sci. USA 112, 1553–1558 (2015)
Chen, Y.-C. et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15, 348–360 (2012)
Baradaran, R., Berrisford, J. M., Minhas, G. S. & Sazanov, L. A. Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013)
Zickermann, V. et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49 (2015)
Letts, J. A. & Sazanov, L. A. Gaining mass: the structure of respiratory complex I — from bacterial towards mitochondrial versions. Curr. Opin. Struct. Biol. 33, 135–145 (2015)
Fiedorczuk, K. et al. Structure of the entire mammalian mitochondrial complex I. Nature http://dx.doi.org/10.1038/nature19794 (2016)
Stroh, A. et al. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279, 5000–5007 (2004)
Yip, C.-Y., Harbour, M. E., Jayawardena, K., Fearnley, I. M. & Sazanov, L. A. Evolution of respiratory complex I: “supernumerary” subunits are present in the alpha-proteobacterial enzyme. J. Biol. Chem. 286, 5023–5033 (2011)
Nübel, E., Wittig, I., Kerscher, S., Brandt, U. & Schägger, H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics 9, 2408–2418 (2009)
Hayashi, H. et al. HIG1, a novel regulator of mitochondrial γ-secretase, maintains normal mitochondrial function. FASEB J. 26, 2306–2317 (2012)
Segade, F., Hurlé, B., Claudio, E., Ramos, S. & Lazo, P. S. Identification of an additional member of the cytochrome c oxidase subunit VIIa family of proteins. J. Biol. Chem. 271, 12343–12349 (1996)
Hüttemann, D. et al. Mice deleted for heart-type cytochrome c oxidase subunit 7a1 develop dilated cardiomyopathy. Mitochondrion 12, 294–301 (2012)
Tsukihara, T. et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl Acad. Sci. USA 100, 15304–15309 (2003)
Swierczek, M. et al. An electronic bus bar lies in the core of cytochrome bc1 . Science 329, 451–454 (2010)
Zhang, Z. et al. Electron transfer by domain movement in cytochrome bc1 . Nature 392, 677–684 (1998)
Shinzawa-Itoh, K. et al. Purification of active respiratory supercomplex from bovine heart mitochondria enables functional studies. J. Biol. Chem. 291, 4178–4184 (2016)
Gao, X. et al. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry 42, 9067–9080 (2003)
Smith, A. L. Preparation, properties, and conditions for assay of mitochondria: slaughterhouse material, small-scale. Methods Enzymol. 10, 81–86 (1967)
Dudkina, N. V., Eubel, H., Keegstra, W., Boekema, E. J. & Braun, H.-P. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl Acad. Sci. USA 102, 3225–3229 (2005)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Scheres, S. H. W. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3, e03665 (2014)
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nature Methods 9, 853–854 (2012)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2013)
Iwata, S. et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71 (1998)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 (2008)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Sazanov, L. A. & Hinchliffe, P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 (2006)
Efremov, R. G. & Sazanov, L. A. Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011)
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999)
Buchan, D. W. A., Minneci, F., Nugent, T. C. O., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41, W349–W57 (2013)
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858 (2015)
Brockmann, C. et al. The oxidized subunit B8 from human complex I adopts a thioredoxin fold. Structure 12, 1645–1654 (2004)
Parris, K. D. et al. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure 8, 883–895 (2000)
Szklarczyk, R. et al. NDUFB7 and NDUFA8 are located at the intermembrane surface of complex I. FEBS Lett. 585, 737–743 (2011)
Zhu, J. et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc. Natl Acad. Sci. USA 112, 12087–12092 (2015)
Ogilvie, I., Kennaway, N. G. & Shoubridge, E. A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 115, 2784–2792 (2005)
Afonine, P. V., Headd, J. J., Terwilliger, T. C. & Adams, P. D. New tool: phenix.real_space_refine. Computational Crystallography Newsletter 4, 43–44 (2013)
Van Kuilenburg, A. B., Van Beeumen, J. J., Van der Meer, N. M. & Muijsers, A. O. Subunits VIIa,b,c of human cytochrome c oxidase. Identification of both ‘heart-type’ and ‘liver-type’ isoforms of subunit VIIa in human heart. Eur. J. Biochem. 203, 193–199 (1992)
Acknowledgements
We thank the MRC LMB Cambridge for the use of the Titan Krios microscope. Data processing was performed using the IST high-performance computer cluster. J.A.L. holds a long-term fellowship from FEBS. K.F. is partially funded by a MRC UK PhD fellowship.
Author information
Authors and Affiliations
Contributions
J.A.L. purified supercomplexes, prepared cryo-EM grids, processed and analysed data, built models and co-wrote the manuscript; K.F. prepared cryo-EM grids, collected cryo-EM data and aided with model building; L.A.S. designed and supervised the project, analysed data and co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks A. Engel, D. Winge and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Supercomplex preparation and image processing procedures.
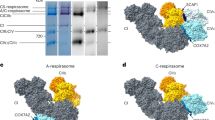
a, BN–PAGE of fractions from a typical sucrose gradient of digitonin extracted supercomplexes. Partially purified CI and molecular weight standards (thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 240 kDa) were also included in the gel. Fraction eight (marked with an asterisk) was taken and used for subsequent supercomplex grid preparation. See the Supplementary Information for full uncropped scans of the gels. b, Representative micrograph of the 1,608 micrographs collected with slightly higher than average particle count; additionally, each micrograph differed slightly in ice thickness and defocus. Scale bar, 100 nm. c, Representative 2D class averages obtained from reference-free classification. d, Classification and refinement procedures used in this study.
Extended Data Figure 2 Local resolution and overall resolutions of supercomplex maps.
a–c, Local resolution estimation by Resmap53 of the tight respirasome (a), the loose respirasome (b) and CICIII2 (c). The supercomplexes are shown from the CI side (left) and the CIII side (right), with the full map (detergent micelle visible), and with the detergent micelle cut out by carving the maps around the protein model (middle images). d, Gold-standard FSC curves for the overall maps. e, f, Per-frame B-factors (e) and Guinier plot intercepts (Cf) (f) used for particle polishing in RELION46 calculated using all good particles.
Extended Data Figure 3 Tight respirasome models and maps for each complex.
Two views of each respirasome complex model fit into the cryo-EM density, with CI in blue (top), CIII in green (middle) and CIV in pink (bottom). The detergent micelle density has been removed for clarity.
Extended Data Figure 4 Loose respirasome models and maps for each complex.
Two views of each respirasome complex model fit into the cryo-EM density, with CI in blue (top), CIII in green (middle) and CIV in pink (bottom). The detergent micelle density has been removed for clarity. The CIV map has been filtered at 8 Å.
Extended Data Figure 5 Supercomplex CICIII2 models and maps for each complex.
Two views of each CICIII2 complex model fit into the cryo-EM density, with CI in blue (top) and CIII in green (bottom). The detergent micelle density has been removed for clarity.
Extended Data Figure 6 Uncut cryo-EM density for the tight respirasome at different contour levels.
Density within 5 Å of the poly-alanine models was coloured with CI in blue, CIII in green and CIV in pink, excess density is coloured grey. a, Low-contour level showing protein and the detergent micelle which is disordered and so has much weaker, fragmented density with no features. b, Medium contour showing mainly protein density. Note that grey density around complexes III and IV is completely featureless, indicating that the known structures account for all observed protein density. c, High contour showing FeS clusters and haem co-factors. Density for N1a cluster in CI, the FeS clusters of UQCRFS1 in CIII and the binuclear copper centre in CIV are not visible due to disorder. N1b cluster is occluded by N4 at this angle. In the loose respirasome, maps peaks for haem a and a3 are not visible in CIV due to disorder.
Extended Data Figure 7 CI model and density.
a–d, Four views of CI showing the density (top) and model (bottom) from within the membrane plane along the side of the membrane arm (a, b), within the membrane plane from the interface of the hydrophilic and hydrophobic arm (c) and from the intermembrane space side (d). a–c, Images are shown with the matrix up and the intermembrane space down. Subunits and density are coloured as indicated. Subunits within the specified colours are: (1) core subunits 75 kDa, 49 kDa, 30 kDa, PSST, TYKY, 51 kDa, 24 kDa, ND1, ND2, ND3, ND4, ND4L, ND5, ND6; (2) supernumerary subunits 39 kDa, B8, B13, 42 kDa, B16.6, B14.7, SDAP-α/β; (3) core subunits 75 kDa and 30 kDa, extended C termini. Supernumerary subunits rebuilt or extended B14, 18 kDa, 13 kDa, B16.6, PGIV, 15 kDa, B14.5b, 42 kDa, ESSS, B22; (4) B17.2, PDSW, B18; (5 and 6) KFYI, B15; (7 and 8) MWFE, B9, B14.5a, MNLL, AGGG, B12, SGDH, B17, ASHI. These subunits have been assigned in our subsequent work32; (9) the unmodelled density, which represents ~7% of the total volume of the CI density. Unmodelled density is primarily located on the solvent-exposed surfaces and represents mainly N- and C termini of known subunits. Similar unmodelled density is also observed in the 5 Å cryo-EM structure of isolated bovine CI (ref. 23).
Extended Data Figure 8 Comparison to previous low-resolution respirasome structures.
a, Models for complexes determined in this study were fit into the cryo-EM density of ref. 18 (a, left) and ref. 19 (a, right), and are consistent with the depictions in those publications, as well as for structures deposited in the PDB. In the structure of the respirasome prepared in amphipols (a, left), all of the complexes are much more separate without forming any of the tight contacts seen here (Fig. 2). This difference may be attributed to the disruptive effects of amphipols or other differences in the preparation or processing. These best fits are compared to our models for the tight (b) and loose (c) respirasomes (both left and right for direct comparison to the models above). For the previous structures in a, 3D-classification was not performed. Therefore, to determine whether the low-resolution maps correspond to an average of the different states observed here, we also fitted our models into a map generated from auto-refinement of all our particles. d, Tight + loose + CICIII2 combined map (Extended Data Fig. 1). This map is dominated by the tight conformation and the model remains distinct from the fits into the previously published low-resolution maps18,19.
Extended Data Figure 9 Modelling of putative higher-order organization of respiratory chain.
One possibility for respiratory strings model could be based on the dimerization of CIV observed in crystal structures24. CIV from the tight respirasome (cyan) was aligned with one of the monomers of CIV from PDB accession code 2X2Q. The second monomer of the crystallographic CIV dimer was then aligned with another copy of respirasome (magenta). a, View from the mitochondrial matrix. b, Side view in the membrane plane. In such an arrangement, there are no clashes between the two copies of the respirasome, with complexes III and IV forming a flat interaction area. The excessive curvature of the model (contrasting with the aligned in plane transmembrane domains in individual respirasomes) may be explained if the contacts between monomers in the crystallographic CIV dimer deviate somewhat from the contacts in the putative CIV dimer within the membrane. Such a symmetric ‘twin’ respirasome may form a building block for putative respiratory strings.
Extended Data Figure 10 COX7a homologues.
a, Sequence alignment of ovine COX7a1, COX7a2 and SCAF1 (also known as COX7a-2RP, COX7a2l). Amino acid changes relative to the COX7a1 sequence are indicated with an asterisk. The N termini of mature COX7a1 and COX7a2 have been determined experimentally69, however, the exact N terminus of mature SCAF1 is unknown. Mass-spectrometry-identified peptides from BN–PAGE indicate that the N terminus contains at least an additional 11 amino acids14, however, these residues are predicted to be unstructured and hence have not been included in the sequence alignment. The sequence numbering is based on mature COX7a1 and full-length protein for SCAF1. b, c, Amino acid differences between COX7a1 and COX7a2 (b) or SCAF1 (c) are mapped onto the structure of bovine COX7a1 indicating that most differences point away from CIV and may contribute to its interactions with other electron transport chain complexes.
Supplementary information
Supplementary Figures
This file contains the uncropped scans with size marker indications for Extended Data Figure 1a. (PDF 421 kb)
Rights and permissions
About this article
Cite this article
Letts, J., Fiedorczuk, K. & Sazanov, L. The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016). https://doi.org/10.1038/nature19774
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19774
This article is cited by
-
SCAF1 drives the compositional diversity of mammalian respirasomes
Nature Structural & Molecular Biology (2024)
-
Di-(2-ethylhexyl) phthalate exposure induces premature testicular senescence by disrupting mitochondrial respiratory chain through STAT5B-mitoSTAT3 in Leydig cell
GeroScience (2024)
-
Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae
Nature Communications (2023)
-
Noncanonical role of singleminded-2s in mitochondrial respiratory chain formation in breast cancer
Experimental & Molecular Medicine (2023)
-
A FRET-based respirasome assembly screen identifies spleen tyrosine kinase as a target to improve muscle mitochondrial respiration and exercise performance in mice
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.