Abstract
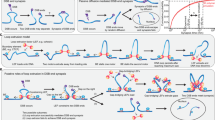
Non-homologous end joining (NHEJ) is the primary pathway for repairing DNA double-strand breaks (DSBs) in mammalian cells1. Such breaks are formed, for example, during gene-segment rearrangements in the adaptive immune system or by cancer therapeutic agents. Although the core components of the NHEJ machinery are known, it has remained difficult to assess the specific roles of these components and the dynamics of bringing and holding the fragments of broken DNA together. The structurally similar XRCC4 and XLF proteins are proposed to assemble as highly dynamic filaments at (or near) DSBs2. Here we show, using dual- and quadruple-trap optical tweezers combined with fluorescence microscopy, how human XRCC4, XLF and XRCC4–XLF complexes interact with DNA in real time. We find that XLF stimulates the binding of XRCC4 to DNA, forming heteromeric complexes that diffuse swiftly along the DNA. Moreover, we find that XRCC4–XLF complexes robustly bridge two independent DNA molecules and that these bridges are able to slide along the DNA. These observations suggest that XRCC4–XLF complexes form mobile sleeve-like structures around DNA that can reconnect the broken ends very rapidly and hold them together. Understanding the dynamics and regulation of this mechanism will lead to clarification of how NHEJ proteins are involved in generating chromosomal translocations3,4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010)
Reid, D. A. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl Acad. Sci. USA 112, E2575–E2584 (2015)
Ghezraoui, H. et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 (2015)
Gelot, C. et al. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell 61, 15–26 (2016)
Cottarel, J. et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 200, 173–186 (2013)
Mani, R. S. et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J. Biol. Chem. 285, 37619–37629 (2010)
Graham, T. G. W., Walter, J. C. & Loparo, J. J. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell 61, 850–858 (2016)
Roy, S. et al. XRCC4’s interaction with XLF is required for coding (but not signal) end joining. Nucleic Acids Res. 40, 1684–1694 (2012)
Roy, S. et al. XRCC4/XLF interaction is variably required for DNA repair and is not required for ligase IV stimulation. Mol. Cell. Biol. 35, 3017–3028 (2015)
Ropars, V. et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA 108, 12663–12668 (2011)
Hammel, M. et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286, 32638–32650 (2011)
Wu, Q. et al. Non-homologous end-joining partners in a helical dance: structural studies of XLF–XRCC4 interactions. Biochem. Soc. Trans. 39, 1387–1392 (2011)
Andres, S. N. et al. A human XRCC4–XLF complex bridges DNA. Nucleic Acids Res. 40, 1868–1878 (2012)
Heller, I., Hoekstra, T. P., King, G. A., Peterman, E. J. & Wuite, G. J. Optical tweezers analysis of DNA-protein complexes. Chem. Rev. 114, 3087–3119 (2014)
Candelli, A., Wuite, G. J. L. & Peterman, E. J. G. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions. Phys. Chem. Chem. Phys. 13, 7263–7272 (2011)
Heller, I. et al. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat. Methods 10, 910–916 (2013)
Hammel, M., Yu, Y., Fang, S., Lees-Miller, S. P. & Tainer, J. A. XLF regulates filament architecture of the XRCC4·ligase IV complex. Structure 18, 1431–1442 (2010)
Mahaney, B. L., Hammel, M., Meek, K., Tainer, J. A. & Lees-Miller, S. P. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol. 91, 31–41 (2013)
Blainey, P. C. et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 16, 1224–1229 (2009)
van Mameren, J. et al. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl Acad. Sci. USA 106, 18231–18236 (2009)
Merkel, R., Nassoy, P., Leung, A., Ritchie, K. & Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 (1999)
Laurens, N. et al. Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat. Commun. 3, 1328 (2012)
Gross, P., Farge, G., Peterman, E. J. G. & Wuite, G. J. L. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 475, 427–453 (2010)
Andres, S. N., Modesti, M., Tsai, C. J., Chu, G. & Junop, M. S. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell 28, 1093–1101 (2007)
Heller, I. et al. Mobility analysis of super-resolved proteins on optically stretched DNA: comparing imaging techniques and parameters. ChemPhysChem 15, 727–733 (2014)
Acknowledgements
We thank P. Fourquet and S. Audebert from the CRCM Proteomics facility for help with MS analysis and G. King and A. S. Biebricher for help with the optical tweezers instruments. This work was supported by VICI grants (G.J.L.W. and E.J.G.P.), a VENI grant (I.H.) and a TopTalent grant (A.C.) from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek, a European Research Council starting grant (G.J.L.W.), the French National Cancer Institute (grant PLBIO13-103) (M.M.), the ARC Foundation for Cancer Research (M.M.), the A*MIDEX project (no. ANR-11-IDEX-0001-02), the ‘Investissements d’Avenir’ French Government program (M.M.), fellowship no. 0558/12-5 from the Brazilian program for Coordination for the Improvement of Higher Education Personnel (A.J.M.), and a fellowship from the Collège of Aix-Marseille Université (H.Z.) The research leading to these results received funding from LASERLABEUROPE (grant agreement no. 284464, the European Commission's Seventh Framework Programme).
Author information
Authors and Affiliations
Contributions
M.M., E.J.G.P. and A.C. conceived the study. I.B., G.S., A.C. and S.J.H. performed the single-molecule experiments. I.H. built the quadruple-trap instrument and advised on the force-fluorescence experiments. A.J.M., H.Z., D.N. and M.M. purified the proteins and performed the biochemical analysis of the protein samples. I.B., G.S., M.M., E.J.G.P. and G.J.L.W. wrote the manuscript. E.J.G.P., M.M. and G.J.L.W. led the research, the analysis and the interpretation of the results. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The optical tweezers and fluorescence technology used in this article are patented and licensed to LUMICKS B.V., in which G.S., A.C., I.H., E.J.G.P. and G.J.L.W. have a financial interest.
Additional information
Reviewer Information
Nature thanks M. Morse, M. Nabuan Naufer, M. Williams and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
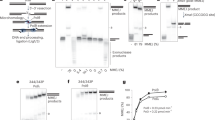
Extended Data Figure 1 Generation of human XLF ATTO-647N-labelled fluorescent variant by the ‘Cys light’ method.
a, Domain architecture of the XLF polypeptide and amino acid sequences of the wild-type and Cys148 variant obtained by site-directed mutagenesis. All Cys residues were changed to Ser except Cys148, leaving a single solvent accessible Cys residue. Three-dimensional model of the XLF dimer where the position–specific labelling sites are indicated by the black arrows. b, Denaturing and reducing polyacrylamide gel electrophoresis of XLF Cys148 variant after labelling with ATTO 647N maleimide. Left, bright-field image of the gel before staining. Centre, the emission of ATTO 647N. Right, is an image of the gel after staining with Coomassie. M, molecular mass markers. c, Mass spectrometry of the labelled full-length protein. Top, the spectra of XLF Cys148 (red) after labelling with ATTO 647N and compared to the wild-type unlabelled protein (black) giving Δm = 790 Da. Bottom, the calculation of the degree of labelling (D.O.L.), as the relative integrated intensity of the labelled protein (dark grey area) versus the unlabelled protein (light grey area) signals, giving 0.82 dye per monomer. d, DNA bridging activity of the XLF variants. Left, a scheme of the bridging assay in which an end-biotinylated 1,000-bp DNA fragment is coupled to streptavidin-coated magnetic beads. Protein-mediated DNA bridging is assessed by recovery of an unlabelled 500-bp DNA fragment. Right, image of an agarose gel stained with ethidium bromide to detect the bridged 500-bp DNA fragment (indicated by the black arrow) and the corresponding quantification. Asterisk indicates biotinylated 1,000-bp DNA fragment adsorbed non-specifically on the surface of the magnetic beads.
Extended Data Figure 2 Generation of human XLF fluorescent variants by eGFP tagging.
a, Domain architecture of the XLF polypeptide and amino acid sequences of the C- and N-terminal eGFP fusions. b, Denaturing and reducing polyacrylamide gel electrophoresis of the purified eGFP fusions. c, DNA bridging activity of the XLF variants. Top, a scheme of the bridging assay where an end-biotinylated 1,000-bp DNA fragment is coupled to streptavidin-coated magnetic beads. Protein-mediated DNA bridging is assessed by recovery of an unlabelled 500-bp DNA fragment. Bottom, an image of an agarose gel stained with ethidium bromide to detect the bridged 500-bp DNA fragment (indicated by the black arrow). Asterisk indicates biotinylated 1,000-bp DNA fragment adsorbed non-specifically on the surface of the magnetic beads.
Extended Data Figure 3 Generation of human XRCC4 fluorescent variants by the ‘Cys light’ method.
a, Domain architecture of the XRCC4 polypeptide and amino acid sequences of the wild-type, C93 and C218 variants obtained by site-directed mutagenesis. All Cys residues were changed to Ala except, respectively C93 and C218, leaving for each variant a single solvent accessible Cys residue. Three-dimensional models of the XRCC4 dimer where the position-specific labelling sites are indicated by the black arrows. b, Denaturing and reducing polyacrylamide gel electrophoresis of XRCC4-C93 and -C218 variants after labelling with Alexa Fluor 555 maleimide. Left, bright-field image of the gel before staining. Centre, the emission of Alexa Fluor 555. Right, an image of the gel after staining with Coomassie. c, Mass spectrometry of the labelled full-length proteins. Top, the spectra of XRCC4-C93 (orange) and XRCC4-C218 (green) after labelling with Alexa Fluor 555 and compared to the wild-type unlabelled protein (black), giving Δm = 830 Da and Δm = 835 Da for labelled XRCC4-C93 and XRCC4-C218, respectively. Bottom, the calculation of the degree of labelling, as the relative integrated intensity of the labelled protein (dark grey area) versus the unlabelled protein (light grey area) signals, giving 0.91 and 0.94 dye per monomer for XRCC4-C93 and XRCC4-C218, respectively. d, DNA bridging activity of the XRCC4 variants. Top, a scheme of the bridging assay in which an end-biotinylated 1,000-bp DNA fragment is coupled to streptavidin-coated magnetic beads. Protein-mediated DNA bridging is assessed by recovery of an unlabelled 500-bp DNA fragment. Bottom, image of agarose gel stained with ethidium bromide to detect the bridged 500-bp DNA fragment (indicated by the black arrow) and the corresponding quantification (the image is from the same gel as in Extended Data Fig. 1d but only the lanes relevant to the analysis of fluorescently labelled XRCC4 are shown). Asterisk indicates biotinylated 1,000-bp DNA fragment adsorbed non-specifically on the surface of the magnetic beads.
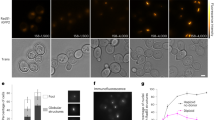
Extended Data Figure 4 Quantification of XRCC4 and XLF binding to dsDNA.
a, Fluorescence image of an overstretched dsDNA molecule in the presence of 100 nM eGFP–XLF. Under the given salt conditions and at this DNA extension, significant DNA melting and formation of ssDNA is expected. XLF shows a high affinity to bind to dsDNA (bright fluorescent signal), whereas it does not appear to bind to ssDNA (dark region). b, c, Size of the DNA-bound protein complexes for XRCC4 (b) and XLF (c) as determined from their fluorescence intensities (after incubation with 50 nM XRCC4 (b) or XLF (c)). d, Sections of a kymograph measured in the protein channel (25 nM eGFP–XLF). Short protein-binding events are visible as brief, local bursts of fluorescence. Events are shorter than the line scan time (10 ms). e, Typical intensity time traces of bound XRCC4 and XLF indicate that the complexes bind as a whole from solution and not monomer by monomer. The decrease of the fluorescence intensity in time is due to photobleaching of the fluorophores. f, Two successive kymographs (separated by white line). Two separate kymographs are recorded owing to technical limitations on the maximum recoding time of XRCC4–Alexa Fluor 555 binding measured using a very low excitation power to reduce the effect of photobleaching. Under these conditions, XRCC4 oligomers stay bound for long periods of time (in the order of several minutes). Scale bars, 5 s and 2 μm. Data are representative examples of 7 (a), 13 (d) and 5 (f) experiments.
Extended Data Figure 5 Properties of protein bridges.
a, b, Bridging also occurs in the absence of free protein in solution. Fluorescence images (XRCC4–Alexa Fluor 555 and XLF–ATTO 647N fluorescence in a, and XRCC4–Alexa Fluor 555 fluorescence in b) acquired in the absence of free protein in solution of two XRCC4–XLF-coated dsDNA molecules before (a) and after (b) wrapping and subsequent unwrapping shows that bridging does not require the presence of free protein in solution. Representative example out of 2 experiments. c, d, DNA–protein bridges also occur in the presence of either XRCC4 or XLF. Fluorescence images of bridges formed by wrapping and subsequent unwrapping in the presence of XRCC4–Alexa Fluor 555 (c) or eGFP–XLF (d). Representative example out of 10 (c) or 19 (d) experiments. e, f, In ~5% of the cases, rupture of protein bridges resulted into two intact dsDNA molecules bound with XRCC4–Alexa Fluor 555 and XLF–ATTO 647N. Fluorescence images before (e) and after (f) rupture of such a protein bridge. Representative example out of 9 experiments. g, h, Fluorescence images taken of an XRCC4–XLF bridge at t = 0 min (g) and t = 95 min (h), showing that the bridge and the protein complexes remain stably bound to the DNA segments. Green, XRCC4; red, XLF, yellow, colocalization. Scale bars, 2 μm.
Extended Data Figure 6 Quantification of diffusion behaviour of XRCC4 and XLF on dsDNA.
a, Typical kymographs showing switching of XLF–ATTO 647N (red signal, top) XRCC4–Alexa Fluor 555 (green signal, middle) and XLF–XRCC4 (yellow signal, bottom) complexes between static and diffusive states. Scale bars, 1 s and 2 μm. b, Typical MSD curve of an individual XRCC4–Alexa Fluor 555 complex. Inset, kymograph of corresponding complex. Red line denotes linear fit to the first three data points; from the slope, a diffusion coefficient of 0.51 μm2 s−1 is determined. See Methods for details on MSD analysis. c, Diffusion coefficients of mobile XRCC4 and XLF complexes at different salt conditions (low salt: 25 mM KCl, high salt: 160 mM KCl) and DNA tensions. d, Diffusion coefficients of mobile XRCC4 protein complexes as a function of complex size. Grey circles, individual data points; black circles, average of 8 successive data points. e, Comparison of the observed diffusion coefficients of XRCC4 (green), XLF (red) and XRCC4–XLF complexes (yellow) to the expected diffusion coefficients based on the helical diffusion model19 (solid black line). To calculate the quantity on the horizontal axis, a hydrodynamic radius of 7–22 nm was used, based on the inner and outer radii of the XRCC4–XLF filament as proposed previously13. f, Average observed dwell times of protein complexes before switching to a different mode. The analysis is performed on 121 events. The shortest dwell time that could be determined with certainty was 1 s. All error bars denote s.e.m.
Extended Data Figure 7 Rupture events in force-extension curves can be caused by DNA–protein bridges or nonspecific sticking of protein-bound DNA to the trapped microspheres.
a, c, Force-extension curves of dsDNA–XRCC4–XLF complexes (red) after incubation at low tension shows rupture events. Black data sets show force-extension curve of bare dsDNA. b, A single, continuous fluorescence kymograph corresponding to red curve in a. At the indicated time (orange arrow in a and b) a protein bridge suddenly ruptures into multiple smaller protein complexes. d, Fluorescence kymograph corresponding to red curve in c reveals that rupture event corresponds to the rupture of a non-specific interaction between a DNA-bound protein complex and the polystyrene microsphere. Scale bars, 1 s and 2 μm.
Extended Data Figure 8 Schematic representation of dual-DNA configurations used in quadruple-trap experiments.
a–c, Wrapped (a), unwrapped (b) and crossed (c) DNA configurations.
Extended Data Figure 9 Quantification of DNA bridging by XRCC4–XLF.
a, b, Normalized length of DNA segments over time during experiments such as shown in Figs 2 and 3. a, The static bridge in the experiment described in Fig. 2 and Supplementary Video 1. b, The mobile bridge in the experiment described in Fig. 3 and Supplementary Video 2.
Extended Data Figure 10 Protein bridges always contain both XRCC4 and XLF.
a, Typical examples of XRCC4–XLF bridges formed by incubating two wrapped DNA molecules in 200 nM eGFP–XLF and 200 nM XRCC4–Alexa Fluor 555 for 2 min. In all 152 bridges that were analysed, clear colocalization of the proteins at the junction is observed.
Supplementary information
Supplementary Figure
This file contains the original source images for all data obtained by electrophoretic separation. (PDF 187 kb)
Bridging by XRCC4-XLF complexes
Video of XRCC4-Alexa Fluor 555 fluorescence (green) and eGFP-XLF fluorescence (red) of the experiment shown in Figure 2. DNA molecules were wrapped, incubated for 2 minutes in a buffer containing 200 nM XRCC4 and 200 nM XLF, brought into a protein-free buffer and subsequently unwrapped. Then individual microspheres were moved to exert tension on the XRCC4-XLF bridge and the DNA ends detach from the microspheres. (MP4 12969 kb)
Mobility of XRCC4-XLF bridges
Video of eGFP-XLF fluorescence bound to two optically trapped DNA molecules bridged by XLF-XRCC4 (individual frames are shown in Figure 3). The movie shows that the bridge slides along one of the DNA molecules when the other DNA molecule is moved. The bridge was formed by incubating the construct for 2 minutes in a buffer containing 200 nM XRCC4 and 200 nM XLF and brought into a protein-free buffer before imaging. (MP4 3292 kb)
XRCC4-XLF complexes keep DNA fragments together after DSBs
Video of XRCC4-Alexa Fluor 555 (green) and eGFP-XLF fluorescence (red) during induction of DSBs in both DNA molecules in an experiment such as shown in Figure 4a-d. A complex of XRCC4 and XLF holds the DNA fragments together in a mobile manner after the occurrence of the DSBs. (MP4 35132 kb)
Rights and permissions
About this article
Cite this article
Brouwer, I., Sitters, G., Candelli, A. et al. Sliding sleeves of XRCC4–XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 (2016). https://doi.org/10.1038/nature18643
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18643
This article is cited by
-
Extracellular vesicle-packaged circBIRC6 from cancer-associated fibroblasts induce platinum resistance via SUMOylation modulation in pancreatic cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
Systematic analysis identifies XRCC4 as a potential immunological and prognostic biomarker associated with pan-cancer
BMC Bioinformatics (2023)
-
Assembly mechanism of the inflammasome sensor AIM2 revealed by single molecule analysis
Nature Communications (2023)
-
DNA double-strand break end synapsis by DNA loop extrusion
Nature Communications (2023)
-
Autopolyploidization affects transcript patterns and gene targeting frequencies in Physcomitrella
Plant Cell Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.