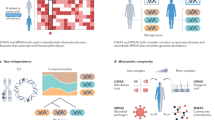
Abstract
When most people think of human development, they tend to consider only human cells and organs. Yet there is another facet that involves human-associated microbial communities. A microbial perspective of human development provides opportunities to refine our definitions of healthy prenatal and postnatal growth and to develop innovative strategies for disease prevention and treatment. Given the dramatic changes in lifestyles and disease patterns that are occurring with globalization, we issue a call for the establishment of 'human microbial observatories' designed to examine microbial community development in birth cohorts representing populations with diverse anthropological characteristics, including those undergoing rapid change.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sender, R., Fuchs, S. & Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Gordon, J., Knowlton, N., Relman, D. A., Rohwer, F. & Youle, M. Superorganisms and holobionts. Microbe 8, 152–153 (2013).
Levison, M. E., Corman, L. C., Carrington, E. R. & Kaye, D. Quantitative microflora of the vagina. Am. J. Obstet. Gynecol. 127, 80–85 (1977).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108 (suppl. 1), 4680–4687 (2011).
Dareng, E. O. et al. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol. Infect. 144, 123–137 (2016).
Koren, O. et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9, e1002863 (2013).
Hillier, S. L. et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N. Engl. J. Med. 333, 1737–1742 (1995).
Horner-Devine, M. C. & Bohannan, B. J. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87, S100–S108 (2006).
Stowell, S. R. et al. Microbial glycan microarrays define key features of host–microbial interactions. Nature Chem. Biol. 10, 470–476 (2014).
Romero, R. et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2, 18 (2014).
Romero, R. et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2, 4 (2014).
DiGiulio, D. B. et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl Acad. Sci. USA 112, 11060–11065 (2015). This study showed that the composition of the vaginal microbiota early in pregnancy may predict subsequent premature birth, which raises questions about how this community of microbes shapes maternal health and pregnancy outcomes.
Aagaard, K. et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 7, e36466 (2012).
Wilson, C. S. Nutritionally beneficial cultural practices. World Rev. Nutr. Diet. 45, 68–96 (1985).
Bisanz, J. E. et al. Microbiota at multiple body sites during pregnancy in a rural Tanzanian population and effects of moringa-supplemented probiotic yogurt. Appl. Environ. Microbiol. 81, 4965–4975 (2015).
MacIntyre, D. A. et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 5, 8988 (2015).
Huang, Y. E. et al. Homogeneity of the vaginal microbiome at the cervix, posterior fornix, and vaginal canal in pregnant Chinese women. Microb. Ecol. 69, 407–414 (2015).
Burke, B. S. & Stevenson, S. S. Nutrition studies during pregnancy; relation of maternal nutrition to condition of infant at birth; study of siblings. J. Nutr. 38, 453–467 (1949).
Koren, O. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480 (2012).
Liu, B. et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE 7, e37919 (2012).
Duran-Pinedo, A. E. et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8, 1659–1672 (2014).
Siqueira, F. M. et al. Intrauterine growth restriction, low birth weight, and preterm birth: adverse pregnancy outcomes and their association with maternal periodontitis. J. Periodontol. 78, 2266–2276 (2007).
DiGiulio, D. B. et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE 3, e3056 (2008).
DiGiulio, D. B. et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 64, 38–57 (2010).
Han, Y. W. et al. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J. Clin. Microbiol. 44, 1475–1483 (2006).
Han, Y. W., Shen, T., Chung, P., Buhimschi, I. A. & Buhimschi, C. S. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J. Clin. Microbiol. 47, 38–47 (2009).
Swati, P., Thomas, B., Vahab, S. A., Kapaettu, S. & Kushtagi, P. Simultaneous detection of periodontal pathogens in subgingival plaque and placenta of women with hypertension in pregnancy. Arch. Gynecol. Obstet. 285, 613–619 (2012).
Costalonga, M. & Herzberg, M. C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162, 22–38 (2014).
Teng, F. et al. Prediction of early childhood caries via spatial–temporal variations of oral microbiota. Cell Host Microbe 18, 296–306 (2015).
Kustner, O. Beitrag zur Lehre von der puerperalen Infection der Neugeborenen. Arch. Gynakol. 11, 256–263 (1877).
Harris, J. W. & Brown, J. H. The bacterial content of the uterus at cesarean section. Am. J. Obstet. Gynecol. 13, 133–143 (1927).
Benirschke, K. Routes and types of infection in the fetus and the newborn. AMA J. Dis. Child. 99, 714–721 (1960).
Verstraelen, H. et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1–2 region of the 16S rRNA gene. PeerJ 4, e1602 (2016).
Mitchell, C. M. et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am. J. Obstet. Gynecol. 212, 611.e1–611.e9 (2015).
Stout, M. J. et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am. J. Obstet. Gynecol. 208, 226.e1–226.e7 (2013).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Bhola, K. et al. Placental cultures in the era of peripartum antibiotic use. Aust. N. Z. J. Obstet. Gynaecol. 48, 179–184 (2008).
Kliman, H. J. Comment on “The placenta harbors a unique microbiome”. Sci. Transl. Med. 6, 254le4 (2014).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Prince, A. L. et al. The placental microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am. J. Obstet. Gynecol. 214, 627.e1–627.e16 (2016).
Kim, M. J. et al. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intraamniotic infection. Lab. Invest. 89, 924–936 (2009).
Hansen, R. et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS ONE 10, e0133320 (2015).
Ardissone, A. N. et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE 9, e90784 (2014).
Romero, R. et al. The role of infection in preterm labour and delivery. Paediatr. Perinat. Epidemiol. 15 (suppl. 2), 41–56 (2001).
Bearfield, C., Davenport, E. S., Sivapathasundaram, V. & Allaker, R. P. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG 109, 527–533 (2002).
Menon, R., Peltier, M. R., Eckardt, J. & Fortunato, S. J. Diversity in cytokine response to bacteria associated with preterm birth by fetal membranes. Am. J. Obstet. Gynecol. 201, 306.e1–306.e6 (2009).
Gomez de Agüero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016). A germ-free mouse model of transient microbial colonization demonstrates that exposure of the mother to microbes during pregnancy shapes immunological development and function in the neonate.
Muglia, L. J. & Katz, M. The enigma of spontaneous preterm birth. N. Engl. J. Med. 362, 529–535 (2010).
McGuire, M. K. & McGuire, M. A. Human milk: mother nature's prototypical probiotic food? Adv. Nutr. 6, 112–123 (2015).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Wu, S., Tao, N., German, J. B., Grimm, R. & Lebrilla, C. B. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 9, 4138–4151 (2010).
Wu, S., Grimm, R., German, J. B. & Lebrilla, C. B. Annotation and structural analysis of sialylated human milk oligosaccharides. J. Proteome Res. 10, 856–868 (2011).
Kunz, C. & Rudloff, S. Biological functions of oligosaccharides in human milk. Acta Paediatr. 82, 903–912 (1993).
Ninonuevo, M. R. et al. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54, 7471–7480 (2006).
Totten, S. M. et al. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J. Proteome Res. 11, 6124–6133 (2012).
Bode, L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22, 1147–1162 (2012).
Thurl, S. et al. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 104, 1261–1271 (2010).
Totten, S. M. et al. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Anal. Bioanal. Chem. 406, 7925–7935 (2014). This paper highlights nanoflow liquid chromatography mass spectrometry, a method that allows the rapid and reproducible detection of HMOs in low-volume biological samples, enabling large-scale clinical studies.
Coppa, G. V. et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics 91, 637–641 (1993).
Niñonuevo, M. R. et al. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J. Agric. Food Chem. 56, 618–626 (2008).
Chaturvedi, P. et al. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11, 365–372 (2001).
De Leoz, M. L. et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J. Proteome Res. 11, 4662–4672 (2012).
Charbonneau, M. R. et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164, 859–871 (2016). Gnotobiotic mouse and piglet models were used to show that sialylated milk oligosaccharides play a causal, microbiota-dependent role in lean body-mass gain, bone growth and metabolism.
Gal, B. et al. Development changes in UDP-N-acetylglucosamine 2-epimerase activity of rat and guinea-pig liver. Comp. Biochem. Physiol. B 108, 13–15 (1997).
Wang, B. Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 (2009).
Wang, B. & Brand-Miller, J. The role and potential of sialic acid in human nutrition. Eur. J. Clin. Nutr. 57, 1351–1369 (2003).
Wang, B. et al. Dietary sialic acid supplementation improves learning and memory in piglets. Am. J. Clin. Nutr. 85, 561–569 (2007).
Yonekawa, T. et al. Sialyllactose ameliorates myopathic phenotypes in symptomatic GNE myopathy model mice. Brain 137, 2670–2679 (2014).
Chen, X. Human milk oligosaccharides (HMOS): structure, function, and enzyme-catalyzed synthesis. Adv. Carbohydr. Chem. Biochem. 72, 113–190 (2015).
Aldredge, D. L. et al. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology 23, 664–676 (2013).
Sundekilde, U. K. et al. Natural variability in bovine milk oligosaccharides from Danish Jersey and Holstein-Friesian breeds. J. Agric. Food Chem. 60, 6188–6196 (2012).
Muoio, D. M. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell 159, 1253–1262 (2014).
Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z. & Dominguez-Bello, M. G. The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117 (2015).
Lewis, Z. T. et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3, 13 (2015).
Huda, M. N. et al. Stool microbiota and vaccine responses of infants. Pediatrics 134, e362–e372 (2014).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014). This study used a machine-learning approach to define normal microbiota development in Bangladeshi infants and children and revealed a persistent defect in microbiota development in children that exhibit undernutrition.
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Sugahara, H., Odamaki, T., Hashikura, N., Abe, F. & Xiao, J. Z. Differences in folate production by bifidobacteria of different origins. Biosci. Microbiota Food Health 34, 87–93 (2015).
Garrido, D., Dallas, D. C. & Mills, D. A. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 159, 649–664 (2013).
Garrido, D. et al. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci. Rep. 5, 13517 (2015).
Marcobal, A. et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10, 507–514 (2011).
De Leoz, M. L. et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J. Proteome Res. 14, 491–502 (2015).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013).
Frese, S. A. & Mills, D. A. Should infants cry over spilled milk? Fecal glycomics as an indicator of a healthy infant gut microbiome. J. Pediatr. Gastroenterol. Nutr. 60, 695 (2015).
Frese, S. A., Parker, K., Calvert, C. C. & Mills, D. A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 3, 28 (2015).
Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015).
Blanton, L. V. et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016).
Schwarzer, M. et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854–857 (2016).
Kau, A. L. et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 7, 276ra24 (2015).
Planer, J. D et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Dogra, S. et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6, e02419-14 (2015).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Arrieta, M. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152 (2015).
Goyal, M. S., Venkatesh, S., Milbrandt, J., Gordon, J. I. & Raichle, M. E. Feeding the brain and nurturing the mind: linking nutrition and the gut microbiota to brain development. Proc. Natl Acad. Sci. USA 112, 14105–14112 (2015).
Levine, M. M., Kotloff, K. L., Nataro, J. P. & Muhsen, K. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin. Infect. Dis. 55 (suppl. 4), S215–S224 (2012).
MAL-ED Network Investigators. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin. Infect. Dis. 59 (suppl. 4), S193–S206 (2014). This paper describes a large, multi-site birth cohort study that includes an effort to serially sample microbial communities in infants to identify correlations between the composition and the development of the microbiota, postnatal growth phenotypes and other facets of health.
Ngure, F. M. et al. Water, sanitation, and hygiene (WASH), environmental enteropathy, nutrition, and early child development: making the links. Ann. NY Acad. Sci. 1308, 118–128 (2014).
Acknowledgements
Work cited from the authors' laboratories was supported in part by grants from the US National Institutes of Health (DK30292, HD061923, AT007079 and AT008759), the Bill & Melinda Gates Foundation, the March of Dimes Foundation and the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.I.G. is co-founder of Matatu, a company that is characterizing the role of diet-by-microbiota interactions in animal health. On completion of his PhD studies, M.R.C. joined Matatu as a research scientist. C.B.L and D.A.M. are co-founders of Evolve Biosystems, a company that is focused on the diet-based manipulation of the gut microbiota. D.A.R. is a member of the scientific advisory board of Seres Health. L.V.B and D.B.D. declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com.reprints.
Rights and permissions
About this article
Cite this article
Charbonneau, M., Blanton, L., DiGiulio, D. et al. A microbial perspective of human developmental biology. Nature 535, 48–55 (2016). https://doi.org/10.1038/nature18845
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18845
This article is cited by
-
A citizen-science-enabled catalogue of the vaginal microbiome and associated factors
Nature Microbiology (2023)
-
Effect of a High-Fat Diet and Probiotic Supplementation on the Gut Microbiota of Maternal Mice at Term Pregnancy and Offspring at Three-Week Postpartum
Current Microbiology (2023)
-
Natural selection for imprecise vertical transmission in host–microbiota systems
Nature Ecology & Evolution (2021)
-
Microbes exploit death-induced nutrient release by gut epithelial cells
Nature (2021)
-
Mediterranean Diet In Healthy Aging
The Journal of nutrition, health and aging (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.