Abstract
Accurate chromosome segregation requires timely dissolution of chromosome cohesion after chromosomes are properly attached to the mitotic spindle. Separase is absolutely essential for cohesion dissolution in organisms from yeast to man1,2. It cleaves the kleisin subunit of cohesin and opens the cohesin ring to allow chromosome segregation. Cohesin cleavage is spatiotemporally controlled by separase-associated regulatory proteins, including the inhibitory chaperone securin3,4,5,6, and by phosphorylation of both the enzyme and substrates7,8,9,10,11,12. Dysregulation of this process causes chromosome missegregation and aneuploidy, contributing to cancer and birth defects. Despite its essential functions, atomic structures of separase have not been determined. Here we report crystal structures of the separase protease domain from the thermophilic fungus Chaetomium thermophilum, alone or covalently bound to unphosphorylated and phosphorylated inhibitory peptides derived from a cohesin cleavage site. These structures reveal how separase recognizes cohesin and how cohesin phosphorylation by polo-like kinase 1 (Plk1) enhances cleavage. Consistent with a previous cellular study13, mutating two securin residues in a conserved motif that partly matches the separase cleavage consensus converts securin from a separase inhibitor to a substrate. Our study establishes atomic mechanisms of substrate cleavage by separase and suggests competitive inhibition by securin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Uhlmann, F., Wernic, D., Poupart, M. A., Koonin, E. V. & Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386 (2000)
Hauf, S., Waizenegger, I. C. & Peters, J. M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323 (2001)
Hornig, N. C., Knowles, P. P., McDonald, N. Q. & Uhlmann, F. The dual mechanism of separase regulation by securin. Curr. Biol. 12, 973–982 (2002)
Ciosk, R. et al. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067–1076 (1998)
Zou, H., McGarry, T. J., Bernal, T. & Kirschner, M. W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–422 (1999)
Waizenegger, I., Giménez-Abián, J. F., Wernic, D. & Peters, J. M. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12, 1368–1378 (2002)
Stemmann, O., Zou, H., Gerber, S. A., Gygi, S. P. & Kirschner, M. W. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 (2001)
Gorr, I. H., Boos, D. & Stemmann, O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol. Cell 19, 135–141 (2005)
Hellmuth, S. et al. Human chromosome segregation involves multi-layered regulation of separase by the peptidyl-prolyl-isomerase Pin1. Mol. Cell 58, 495–506 (2015)
Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M. A. & Nasmyth, K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472 (2001)
Hauf, S. et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 (2005)
Katis, V. L. et al. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell 18, 397–409 (2010)
Nagao, K. & Yanagida, M. Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes Cells 11, 247–260 (2006)
Viadiu, H., Stemmann, O., Kirschner, M. W. & Walz, T. Domain structure of separase and its binding to securin as determined by EM. Nature Struct. Mol. Biol. 12, 552–553 (2005)
Gligoris, T. G. et al. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science 346, 963–967 (2014)
Huis in ’t Veld, P. J. et al. Characterization of a DNA exit gate in the human cohesin ring. Science 346, 968–972 (2014)
Sullivan, M., Lehane, C. & Uhlmann, F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nature Cell Biol. 3, 771–777 (2001)
Matsuo, K. et al. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22, 915–921 (2012)
Renatus, M., Stennicke, H. R., Scott, F. L., Liddington, R. C. & Salvesen, G. S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl Acad. Sci. USA 98, 14250–14255 (2001)
Chai, J. et al. Structural basis of caspase-7 inhibition by XIAP. Cell 104, 769–780 (2001)
Srinivasula, S. M., Ahmad, M., Fernandes-Alnemri, T. & Alnemri, E. S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1, 949–957 (1998)
Shi, Y. Caspase activation: revisiting the induced proximity model. Cell 117, 855–858 (2004)
Kitajima, T. S. et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441, 46–52 (2006)
Riedel, C. G. et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441, 53–61 (2006)
Tang, Z. et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell 10, 575–585 (2006)
Ishiguro, T., Tanaka, K., Sakuno, T. & Watanabe, Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nature Cell Biol. 12, 500–506 (2010)
Liu, H., Rankin, S. & Yu, H. Phosphorylation-enabled binding of SGO1–PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nature Cell Biol. 15, 40–49 (2013)
Sullivan, M., Hornig, N. C., Porstmann, T. & Uhlmann, F. Studies on substrate recognition by the budding yeast separase. J. Biol. Chem. 279, 1191–1196 (2004)
Zhang, N. et al. Overexpression of separase induces aneuploidy and mammary tumorigenesis. Proc. Natl Acad. Sci. USA 105, 13033–13038 (2008)
Sun, Y. et al. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell 137, 123–132 (2009)
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Lebedev, A. A. et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D 68, 431–440 (2012)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Acknowledgements
We thank D. Rosenbaum for the C. thermophilum complementary DNA, H. Ball for peptide synthesis, and D. Tomchick and Z. Chen for assistance with data collection. Diffraction data of the selenomethionine separase were collected at the Advanced Light Source at Lawrence Berkeley National Laboratory with the help of its staff. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract number DE-AC02-05CH11231. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work is supported by the Cancer Prevention and Research Institute of Texas (RP110465-P3 to H.Y.), the National Institutes of Health (GM107415 to X.L.), and the Welch Foundation (I-1441 to H.Y.). H.Y. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Z.L. performed all experiments in this study with advice from H.Y. X.L. provided assistance with structure refinement. Z.L. and H.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Sequence alignment of the SPDs from multiple species.
The alignment is generated using the online ESPript 2.0 server. Secondary structural elements of ctSPD are indicated above the sequences, with the same labelling and colour schemes as in Fig. 1d (PPD, blue; APD, green; the helical insert in PPD, cyan). Abbreviations: ct, Chaetomium thermophilum; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe; xt, Xenopus tropicalis; hs, Homo sapiens.
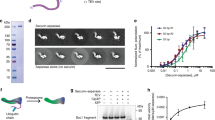
Extended Data Figure 2 Purification, activity, inhibition, and autocleavage of active ctSPD.
a, Coomassie-stained gel of purified recombinant ctSPD wild type (WT) and C2110S. b, Autoradiograph of the ctSPD cleavage assay with 35S-ctScc1 as substrate. c, Chemical structure of the acyloxymethyl ketone (AMK) inhibitor derived from the ctScc1 cleavage site. d, Autoradiograph of the ctSPD cleavage assay with 35S-ctScc1 as substrate, in the absence or presence of increasing doses of the AMK inhibitor depicted in c. e, Coomassie-stained SDS–PAGE gel of purified recombinant ctSPD WT or C2110S treated with the indicated doses of the ctScc1-AMK peptide inhibitor. The positions of unmodified ctSPD and ctSPD–inhibitor conjugates are indicated. f, Coomassie-stained gel of recombinant ctSPD1632–2223 WT or non-cleavable (NC) mutant. The ctSPDNC mutant contains the E1643R and R1646E mutations. The positions of intact and autocleaved ctSPD proteins are indicated.
Extended Data Figure 3 Comparison between the folding topologies of ctSPD and the caspase 9 dimer (Protein Data Bank accession number 1JXQ).
The labelling and colour schemes are the same as in Fig. 1d, e. H, the catalytic histidine; C, the catalytic cysteine.
Extended Data Figure 4 Contributions of the L4 loop and a surface pocket to the protease activity of ctSPD.
a, Cartoon of the crystal structure of ctSPD, with the PPD coloured blue, the APD coloured green, the helical insert in PPD coloured cyan, and an N-terminal tag peptide coloured yellow. The N and C termini are indicated. All secondary structure elements are labelled. Loops with no visible electron densities are indicated by dashed lines. Loop 4 (L4) is coloured magenta. H2083 and C2110 of the catalytic dyad are shown as sticks. The orientation of ctSPD in this figure is related to that in Fig. 1d by a 180° rotation along the vertical axis. c, Representative autoradiograph of the 35S-ctScc1 cleavage assay by WT ctSPD or the indicated mutants. Bottom: Coomassie-stained gel of ctSPD proteins used in the assay. Quantification of the relative protease activities of ctSPD WT and mutants is shown in Fig. 2b. The protease activity is defined as the ratio between intensities of the two major ctScc1 cleavage products and that of the uncleaved ctScc1. c, Cartoon of the crystal structure of ctSPD, in the same orientation as in Fig. 1d. The tag peptide (HSQLEVLFQGP) is shown as sticks, overlaid with its 2Fo − Fc electron density map contoured at 1.0σ. d, Representative autoradiograph of the 35S-ctScc1 cleavage assay by WT ctSPD or the indicated mutants. Bottom: Coomassie-stained gel of ctSPD proteins used in the assay. Quantification of the relative protease activities of ctSPD WT and mutants is shown in Fig. 2d.
Extended Data Figure 5 Interactions between the helical insert and the APD.
a, b, Zoomed-in views of cartoons of ctSPD in two orientations that are related by a 180° rotation along the vertical axis. Residues at the interface between the helical insert of the PPD and the APD are shown in sticks and labelled. c, Coomassie-stained gel of lysates of bacteria expressing the indicated ctSPD mutants and treated without (–) or with (+) isopropyl β-d-1-thiogalactopyranoside (IPTG) and eluates from Ni2+-NTA beads that had been incubated with the IPTG lysates. d, Autoradiograph of the 35S-ctScc1 cleavage assay by WT ctSPD or the indicated mutants. Bottom: Coomassie-stained gel of ctSPD proteins used in the assay.
Extended Data Figure 6 Phospho-regulation and specificity determinants of separase-mediated cohesin cleavage.
a, Autoradiograph of the ctSPD cleavage reactions of 35S-ctScc1 WT or S210A, treated with or without human (hs) Plk1 or its inhibitor BI2536. b, Autoradiograph of the ctSPD cleavage reactions, with 35S-ctScc1 WT or the phospho-mimicking S210E as substrates. c, Zoomed-in view of the cartoon of ctSPD bound covalently to the ctScc1-AMK inhibitor. The catalytic dyad residues C2110 and H2083 are shown as red sticks. The covalently bound inhibitor is shown as yellow sticks, overlaid with its 2Fo − Fc electron density map contoured at 1.0σ. d, Zoomed-in view of the S4 pocket of ctSPD that recognizes the P4 glutamate. Dashed lines indicate hydrogen bonds or favourable electrostatic interactions. The orange sphere indicates a water molecule. e, Mapping of the aberrant ctScc1 cleavage site by ctSPD D2151A. Top: sequence alignment of the aberrant site of D2151A and the major site of WT. Bottom: autoradiograph of the cleavage reactions of ctSPDWT or ctSPDD2151A with the indicated 35S-ctScc1 proteins as substrates. Asterisk marks the aberrant cleavage product by ctSPDD2151A. f, Charge reversal mutants of ctSPD fail to cleave complementary charge reversal mutants of ctScc1. Autoradiograph of the cleavage assay of WT ctSPD or the indicated mutants, with 35S-ctScc1 WT or mutants as substrates. Bottom: Coomassie-stained gel of ctSPD proteins used in the assay.
Extended Data Figure 7 Conservation of substrate-binding residues in human separase.
a, Two different views of the cartoon of the structure of ctSPD–pAMK (green) and a homology model of human (hs) SPD (magenta). The phospho-AMK peptide is shown in sticks. The homology model of hsSPD was generated with SWISS-MODEL. The coordinates of the model are available upon request. b, c, Zoomed-in views of the S1 and S4 pockets of the hsSPD model.
Extended Data Figure 8 Structural basis of phosphorylation-stimulated Scc1 cleavage.
a, Autoradiograph of the ctSPD cleavage reactions of 35S-ctScc1 WT or I211A, treated with or without hsPlk1. b, Zoomed-in view of the surface drawing of ctSPD–pAMK. The surface is coloured according to the electrostatic potential, with red, blue, and white representing negative, positive, and neutral charges, respectively. The covalently bound peptide is shown as sticks. c, Coomassie-stained gel of the indicated ctSPD proteins used in the assays described in Figs 3f and 4c. d, Quantification of the fold of Plk1 stimulation in ctScc1 cleavage by ctSPD WT and the indicated mutants as described in Fig. 3f. Error bars, s.d. (n = 3 independent experiments).
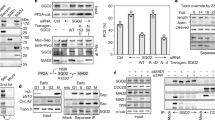
Extended Data Figure 9 Interactions between ctsecurin and ctseparase.
a, Coomassie-stained gel of recombinant ctseparase–ctsecurin complexes and ctSPD expressed in insect cells. FL, full length. b, Autoradiograph of the ctScc1 cleavage reactions by the ctseparase–ctsecurin complexes and ctSPD. c, Autoradiograph of the cleavage reactions of 35S-ctsecurin WT or mutants with or without ctSPD. d, Coomassie-stained gel of recombinant Strep-tagged ctseparase1–1500 or the ctseparase1–1500–ctsecurin complex bound to Strep-Tactin beads. e, Autoradiograph of the ctScc1 cleavage reactions by ctSPD, in the absence or presence of varying concentrations of the ctsecurin153–177 or ctsecurin153–177 3A peptides. The EVE motif is mutated to AAA in the ctsecurin153–177 3A peptide.
Supplementary information
Supplementary Figure 1
This file contains the uncropped gels and autoradiographs for Figures 1c, 2e, 3d, 3f, 4b, 4c, and Extended Data Figures 2a, 2b, 2d, 2e, 2f, 4b, 4d, 5d, 6a, 6b, 6e, 6f, 8a, 8c, 9a, 9b, 9c, 9d, 9e. (PDF 696 kb)
Rights and permissions
About this article
Cite this article
Lin, Z., Luo, X. & Yu, H. Structural basis of cohesin cleavage by separase. Nature 532, 131–134 (2016). https://doi.org/10.1038/nature17402
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17402
This article is cited by
-
Structural basis of human separase regulation by securin and CDK1–cyclin B1
Nature (2021)
-
Two giants of cell division in an oppressive embrace
Nature (2021)
-
A prometaphase mechanism of securin destruction is essential for meiotic progression in mouse oocytes
Nature Communications (2021)
-
Separase-triggered apoptosis enforces minimal length of mitosis
Nature (2020)
-
Securin-independent regulation of separase by checkpoint-induced shugoshin–MAD2
Nature (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.