Abstract
Mutations disabling the TP53 tumour suppressor gene represent the most frequent events in human cancer and typically occur through a two-hit mechanism involving a missense mutation in one allele and a ‘loss of heterozygosity’ deletion encompassing the other. While TP53 missense mutations can also contribute gain-of-function activities that impact tumour progression, it remains unclear whether the deletion event, which frequently includes many genes, impacts tumorigenesis beyond TP53 loss alone. Here we show that somatic heterozygous deletion of mouse chromosome 11B3, a 4-megabase region syntenic to human 17p13.1, produces a greater effect on lymphoma and leukaemia development than Trp53 deletion. Mechanistically, the effect of 11B3 loss on tumorigenesis involves co-deleted genes such as Eif5a and Alox15b (also known as Alox8), the suppression of which cooperates with Trp53 loss to produce more aggressive disease. Our results imply that the selective advantage produced by human chromosome 17p deletion reflects the combined impact of TP53 loss and the reduced dosage of linked tumour suppressor genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
17p13 shRNA library specification is provided in Supplementary Information. The raw and analysed RNA sequence data have been deposited in the Gene Expression Omnibus under accession number GSE69654.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011)
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008)
Xue, W. et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl Acad. Sci. USA 109, 8212–8217 (2012)
Solimini, N. L. et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science 337, 104–109 (2012)
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013)
Miller, L. D. et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl Acad. Sci. USA 102, 13550–13555 (2005)
Petitjean, A., Achatz, M. I., Borresen-Dale, A. L., Hainaut, P. & Olivier, M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 26, 2157–2165 (2007)
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013)
Wattel, E. et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood 84, 3148–3157 (1994)
El-Ghammaz, A. M., Abdelwahed, E., Mostafa, N. N. & Mansour, D. A. De novo deletion 17p13.1 as a predictor for disease progression in chronic lymphocytic leukemia. Clin. Exp. Med. 15, 493–499 (2015)
Scuoppo, C. et al. A tumour suppressor network relying on the polyamine–hypusine axis. Nature 487, 244–248 (2012)
Wales, M. M. et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nature Med. 1, 570–577 (1995)
Ahn, Y. H. et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor γ2 expression. Mol. Cell. Biol. 31, 4270–4285 (2011)
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011)
Adams, D. J. et al. Mutagenic insertion and chromosome engineering resource (MICER). Nature Genet. 36, 867–871 (2004)
Schmitt, C. A. et al. Dissecting p53 tumor suppressor functions in vivo . Cancer Cell 1, 289–298 (2002)
Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 (1999)
Monti, S. et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell 22, 359–372 (2012)
Levine, A. J. & Oren, M. The first 30 years of p53: growing ever more complex. Nature Rev. Cancer 9, 749–758 (2009)
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009)
Olive, K. P. et al. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 119, 847–860 (2004)
Hanel, W. et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 20, 898–909 (2013)
Hemann, M. T. et al. Suppression of tumorigenesis by the p53 target PUMA. Proc. Natl Acad. Sci. USA 101, 9333–9338 (2004)
Kelly, G. L. et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53 . Genes Dev. 28, 58–70 (2014)
Tang, D. G. et al. Suppression of W256 carcinosarcoma cell apoptosis by arachidonic acid and other polyunsaturated fatty acids. Int. J. Cancer 72, 1078–1087 (1997)
Balatti, V. et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 112, 2169–2174 (2015)
Slovak, M. L. et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 96, 4075–4083 (2000)
Byrd, J. C. et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 100, 4325–4336 (2002)
Chen, C. et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 25, 652–665 (2014)
Salaverria, I. et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J. Clin. Oncol. 25, 1216–1222 (2007)
Rubio-Moscardo, F. et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood 105, 4445–4454 (2005)
Chen, W. et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood 107, 2477–2485 (2006)
Bea, S. et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 106, 3183–3190 (2005)
Mestre-Escorihuela, C. et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood 109, 271–280 (2007)
Rücker, F. G. et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 119, 2114–2121 (2012)
Chigrinova, E. et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood 122, 2673–2682 (2013)
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013)
Brown, T. Southern blotting. Curr. Protoc. Prot. Sci. 13, 4G:A.4G.1–4G:A.4G. 8 (2001)
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985)
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003)
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA 93, 5860–5865 (1996)
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994)
Marino, S. & Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000)
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992)
Kamijo, T. et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF . Cell 91, 649–659 (1997)
Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011)
Hemann, M. T. et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436, 807–811 (2005)
Fellmann, C. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports 5, 1704–1713 (2013)
Chicas, A. et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17, 376–387 (2010)
Simpson, E. M. et al. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nature Genet. 16, 19–27 (1997)
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011)
Baslan, T. et al. Genome-wide copy number analysis of single cells. Nature Protocols 7, 1024–1041 (2012)
Baslan, T. & Hicks, J. Single cell sequencing approaches for complex biological systems. Curr. Opin. Genet. Dev. 26, 59–65 (2014)
Folch, J., Lees, M. & Sloane Stanley, G. H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957)
Ye, X. et al. Development and validation of a UPLC-MS/MS method for quantification of SKLB010, an investigational anti-inflammatory compound, and its application to pharmacokinetic studies in beagle dogs. J. Pharm. Biomed. Anal. 56, 366–372 (2011)
Acknowledgements
We thank J. P. Morris, L. Dow, D. Tschaharganeh, E. Manchado, T. Kitzing, E. Loizou and other Lowe laboratory members for their critical discussions and technical help, C. J. Sherr for invaluable advice, L. Dai and M. Tang for liquid chromatography–mass spectrometry support, and Y. Park and S. Kim for their help in constructing the 11B3 mouse model. This work was supported by a programme project and an R01 grant from the National Cancer Institute (S.W.L.), a Center grant for Cancer Target Discovery and Development (S.W.L.), and a Memorial Sloan Kettering Cancer Center Support Grant. Y.L. was supported by an American Association for Cancer Research Millennium Fellowship in Lymphoma Research. C.C. is supported by the Thousand Young Talents Plan and the National Natural Science Foundation of China (81522003, 81570150). S.W.L. is the Geoffrey Beene Chair for Cancer Biology and a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
Y.L. and S.W.L. designed the experiments. Y.L., C.C., S.A., Z.X. and L.C. performed the experiments. Y.L., C.C., Z.X., T.N. and S.W.L. analysed data. Y.L., C.S. and A.A.M. designed the 11B3 model, Y.L., C.S., J.G., B.B., E.R.K., T.B., B.S., T.N., Q.W., N.S. and R.L.L. contributed to the human cancer genomic analysis. Y.L., C.C., C.D.R. and S.W.L. organized data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
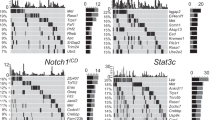
Extended Data Figure 1 The frequency and prognostic impact of chromosome 17p deletion with the copy number loss identified by GISTIC.
a, The ratio of chromosome-copy-number-altered cases within TP53-mutated cases compared to those within wild-type cases. TP53 mutations were statistically correlated with 17p loss (P < 0.001) but also other copy number events (P < 0.001). b, Peaks of copy number loss identified by the GISTIC algorithm in NHL or AML. x axis, GISTIC q value; y axis, chromosome. q < 0.25 is considered as significant. c, Overall survival of human diffuse large B-cell lymphoma (DLBCL) patients with chromosome 17p deletion is significantly shortened compared to those with no 17p copy number variants, as annotated from the Gene Expression Omnibus GSE34171 series. *P < 0.05 (log-rank test).
Extended Data Figure 2 Generation of a chromosome 11B3 conditional knockout mouse.
a, Top, strategy to introduce 5′ HPRT gene and loxP site telomeric to Sco1 on chromosome 11B3 with MICER clone MHPN91j22. Bottom, Southern blot demonstrating correct targeting of the derived ES cells. st, single-targeted allele; wt, wild-type. Blue arrowheads denote loxP sites. b, Top, strategy to introduce 3′ HPRT gene and loxP site centromeric to Alox12 on chromosome 11B3 with MICER clone MHPP248j19. Bottom, Southern blot demonstrating correct targeting of the derived ES cells. dt, double-targeted allele. c, Top, diagram showing the expected PCR results and drug-resistance phenotypes of doubly targeted ES cells harbouring loxP sites in cis versus in trans. GR, G418 (neomycin) resistance; PR, puromycin resistance; HR, HAT resistance. df, deleted allele; dp, duplicated allele. Green bar indicates the PCR product location and length. Bottom, PCR results show different ES cell clones generated in a and b.
Extended Data Figure 3 11B3 recombination and lymphomagenesis in Eu-Myc model.
a, The extent of 11B3 deletion in peripheral blood cells, as determined by semi-quantitative PCR, in 11B3fl/+ mice crossed to Cd19-cre (left), Mx1-cre (middle) or Vav1-cre (right). Genomic DNA from 11B3+/Δ ES cells was mixed with 11B3fl/+ cells at different ratios (5% or 20%) as a standard. For Mx1-cre, 6–8-week-old mice were treated with polyinosinic:polycytidylic acid (poly(I:C)) (15 mg kg−1 every other day, 7 times) by intraperitoneal injection. b, Partial 11B3 deletion in Vav1-cre;11B3fl/+ pre-B cells as determined by semi-quantitative PCR, indicating incomplete recombination. c, Complete Trp53fl/+ recombination in Vav1-cre;Trp53fl/+ pre-B cells as determined by PCR. d, Tumour-free survival of Eμ-Myc;Vav1-cre;Trp53fl/+ (n = 9), Eμ-Myc;Vav1-cre;11B3fl/+ (n = 12) and Eμ-Myc;Vav1-cre (n = 6) mice shows that 11B3-deleted tumours have longer tumour latency than Trp53-loss-only controls. ***P < 0.001 (log-rank test).
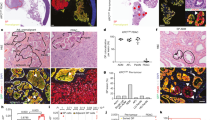
Extended Data Figure 4 Charaterization of 11B3-deleted lymphoma.
a, Immunophenotypes of B220+ Eμ-Myc lymphomas generated from Vav1-cre;p53fl/+ or Vav1-cre;11B3fl/+. 11B3-deleted lymphomas were either IgM−IgD− or IgM+IgD+ while all the Trp53-null lymphomas were IgM−IgD−. b, Haematoxylin and eosin (H&E) stainings of lymph node, spleen and liver of moribund, lymphoma-bearing mice originating from Eμ-Myc;Vav1-cre;11B3fl/+ or Eμ-Myc;Vav1-cre;Trp53fl/+ genotypes. Scale bar, 50 μm. c, 11B3-deleted lymphoma cells isolated from enlarged lymph nodes are more resistant to chemotherapy drugs 4-hydroxycyclophosphamide (left) and vincristine (right), by in vitro drug sensitivity assay. Shown are representative results of three 11B3Δ/Trp53Δframeshift (11B3) or Trp53Δ/Δ (p53) lymphoma cell lines assayed in quadruplicate. *P < 0.05 (Student’s two-tailed t-test). d, e, No functional p53 was detected in various 11B3-deleted tumours as determined by western blotting of p53 and RT–PCR analysis of p21 induction after 4-h ADR treatment. Eμ-Myc;Arf−/− (Trp53+/+) lymphomas were used as a positive control and p21 levels were normalized to untreated cells. Tumours shown in d were identified as missense (tumour 711) or frameshift mutations (tumour 723), while those in e had no detectable mutation. In total eight tumours were analysed. f, The scope of p53 mutations detected in chromosome 11B3-deleted lymphoma cells as determined by sequencing (n = 12). DBD, DNA-binding domain; FS, frameshift mutation; INS, insertion mutation; MS, missense mutation; TAD, transactivation domain; TET, tetramerization domain.
Extended Data Figure 5 Tumours in mice heterozygous for Trp53 mutations lose heterozygosity by duplicating the mutant Trp53 allele.
a, No chromosome 11B3 deletion was detected in various Trp53 heterozygous mutants. Relative allele copy number of various chromosome 11B3 genes, as determined by qPCR analysis of genomic DNA from Eμ-Myc lymphomas derived from germline mice harbouring the following additional alleles: Vav1-cre;Trp53fl/+(exon 2–10 flanked), Trp53+/− (exon 2–6 deleted), Vav1-cre;Trp53LSL-R270H/+ or Vav1-cre;Trp53LSL-R172H/+. Rpa3 on chromosome 6 was used as an endogenous normalization control. b, SNP analysis of tumour or normal tissue (tail) genomic DNA harvested from mice in a, indicating that uniparental disomy occurred during Trp53 LOH, in that C57BL/6 (B6)-derived wild-type Trp53 allele is replaced by 129-derived Trp53 mutant allele. Note that all Trp53-engineered alleles retain 129-derived SNPs; the germline wild-type Trp53 allele is C57BL/6-derived. c, Cartoon summary of the results from a and b.
Extended Data Figure 6 A Trp53 shRNA induces equivalent knockdown in cells with one or two alleles of the Trp53 gene.
Pre-B cells were isolated from Trp53+/+ or Trp53+/− bone marrow, and then transduced with GFP-linked Trp53 shRNA (shp53). GFP+ cells were sorted out by fluorescence-activated cell sorting, and treated with control wild-type pre-B cells in the present of vehicle or 1 μg ml−1 ADR for 4 h. p53 and p21 levels were detected by western blotting and RT–qPCR, respectively. Shown is the representative result of three independent experiments.
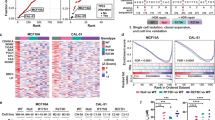
Extended Data Figure 7 In the Eμ-Myc model, Trp53 and Eif5a cooperate in tumorigenesis.
Two-colour assay for the cooperation of Trp53 and Eif5a deficiencies on lymphoma genesis. Eμ-Myc HSPCs retrovirally co-transduced with GFP- (shEif5a or shRen) and mCherry-linked shRNAs (shRen, shp53) were transplanted into sublethally-irradiated syngeneic recipients (n = 5 per group). a, b, The resulting tumours were analysed by flow cytometry (a) and the percentage of GFP+mCherry+ lymphoma cells in each configuration was quantified (b). Error bars represent s.d. *P < 0.05, ***P < 0.001 (two tailed t-test).
Extended Data Figure 8 Alox15b deficiency promotes tumorigenesis and increases AA levels.
a, Enrichment fold of shAlox15b.1252 and shAlox15b.2865 in resulting tumours (Fig. 3i, j) compared to those in initiating shRNA libraries. b, Knockdown efficiency of shAlox15b.1252 and shAlox15b.2865 compared to control shRen in NIH3T3 cells, as detected by western blotting and quantitated by ImageJ. c, Relative levels of AA per cell are increased with Alox15b shRNAs as measured by liquid chromatography–mass spectrometry (LC-MS). NIH3T3 cells were transduced with shRen or shAlox15b. n = 3. **P < 0.01 (unpaired two tailed t-test). d, Relative levels of AA per cell in 11B3Δ/Trp53Δframeshift (11B3) lymphoma cells are higher than control cells with Trp53Δ / Δ (p53) as measured by LC-MS. n = 2. P = 0.056 (unpaired two tailed t-test). e, In vitro AA treatments reduce apoptosis, as measured by annexin V staining of pre-B cells after 20 h treatment of indicated concentration of AA. n = 4. *P < 0.05; ***P < 0.001 (unpaired two tailed t-test).
Extended Data Figure 9 11B3 deletion accelerates leukaemogenesis beyond Trp53 loss alone and decreases sensitivity to the BET-protein inhibitor JQ-1.
a, The percentage of 11B3 deletion as determined by qPCR in premalignant c-Kit+ HSPCs (n = 2) and resulting leukaemia cells (tumour; n = 4). **P < 0.01 (unpaired two-tailed t-test). b, Overall survival of recipient mice transplanted with HSPCs from Vav1-cre;11B3fl/Trp53fl or Vav1-cre;Tr53fl/fl co-transduced with both Nf1 and Mll3 shRNAs. **P < 0.01 (log-rank test). c, Complete blood cell counts of recipient mice indicate that there are more total white blood cells (WBCs) and neutrophils, and fewer red blood cells in Vav1-cre;11B3fl/Trp53fl mice compared with the Vav1-cre;Trp53fl/fl control group at 8 weeks post-transplantation. (Note that two mice from each group died before analysis and were not included.) d, Flow cytometry analysis of GFP+mCherry+ leukaemic cells in the bone marrow of moribund mice in a shows that leukaemia cells are myeloid cells in origin and contain both shNf1 and shMll3. e, f, In vitro drug sensitivity of leukaemia cells to araC (e) and the BET-bromodomain inhibitor JQ-1 (f). Shown are representative results of three 11B3Δ/Trp53Δ and two Trp53Δ/Δ leukaemia cell lines assayed in quadruplicate. *P < 0.05 (Student’s two-tailed t-test).
Supplementary information
Supplementary Information
This file contains a Supplementary Figure, which shows the uncropped blots with size markers for Figure 3b and Extended Data Figures 2a, 2b, 4d, 4e, 6 and 8b, and Supplementary Tables 1-3. (PDF 757 kb)
Supplementary Data
This fie contains Supplementary Data. (XLSX 36 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Chen, C., Xu, Z. et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531, 471–475 (2016). https://doi.org/10.1038/nature17157
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17157
This article is cited by
-
Machine-learning analysis reveals an important role for negative selection in shaping cancer aneuploidy landscapes
Genome Biology (2024)
-
Arachidonic acid inhibit granulosa cell function by affecting metabolic function of liver in brown adipose transplantation rats
Journal of Ovarian Research (2024)
-
Homogenous TP53mut-associated tumor biology across mutation and cancer types revealed by transcriptome analysis
Cell Death Discovery (2023)
-
COX-2/PGE2 upregulation contributes to the chromosome 17p-deleted lymphoma
Oncogenesis (2023)
-
TP53-mutated acute myeloid leukemia and myelodysplastic syndrome: biology, treatment challenges, and upcoming approaches
Annals of Hematology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.