Abstract
Translation of messenger RNAs lacking a stop codon results in the addition of a carboxy-terminal poly-lysine tract to the nascent polypeptide, causing ribosome stalling. Non-stop proteins and other stalled nascent chains are recognized by the ribosome quality control (RQC) machinery and targeted for proteasomal degradation. Failure of this process leads to neurodegeneration by unknown mechanisms. Here we show that deletion of the E3 ubiquitin ligase Ltn1p in yeast, a key RQC component, causes stalled proteins to form detergent-resistant aggregates and inclusions. Aggregation is dependent on a C-terminal alanine/threonine tail that is added to stalled polypeptides by the RQC component, Rqc2p. Formation of inclusions additionally requires the poly-lysine tract present in non-stop proteins. The aggregates sequester multiple cytosolic chaperones and thereby interfere with general protein quality control pathways. These findings can explain the proteotoxicity of ribosome-stalled polypeptides and demonstrate the essential role of the RQC in maintaining proteostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Preissler, S. & Deuerling, E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 (2012)
Pechmann, S., Willmund, F. & Frydman, J. The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 (2013)
Lykke-Andersen, J. & Bennett, E. J. Protecting the proteome: eukaryotic cotranslational quality control pathways. J. Cell Biol. 204, 467–476 (2014)
Frischmeyer, P. A. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002)
van Hoof, A., Frischmeyer, P. A., Dietz, H. C. & Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002)
Barrett, L., Fletcher, S. & Wilton, S. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 69, 3613–3634 (2012)
Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052 (2014)
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014)
Lu, J. & Deutsch, C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86 (2008)
Koutmou, K. S. et al. Ribosomes slide on lysine-encoding homopolymeric A stretches. Elife 4, (2015)
Bengtson, M. H. & Joazeiro, C. A. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010)
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012)
Defenouillere, Q. et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl Acad. Sci. USA 110, 5046–5051 (2013)
Shao, S., von der Malsburg, K. & Hegde, R. S. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648 (2013)
Verma, R., Oania, R. S., Kolawa, N. J. & Deshaies, R. J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2, e00308 (2013)
Shao, S. & Hegde, R. S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell 55, 880–890 (2014)
Lyumkis, D. et al. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl Acad. Sci. USA 111, 15981–15986 (2014)
Shao, S., Brown, A., Santhanam, B. & Hegde, R. S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 (2015)
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015)
Shoemaker, C. J., Eyler, D. E. & Green, R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372 (2010)
Chu, J. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA 106, 2097–2103 (2009)
Krishnan, R. & Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005)
Mukhopadhyay, S., Krishnan, R., Lemke, E. A., Lindquist, S. & Deniz, A. A. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl Acad. Sci. USA 104, 2649–2654 (2007)
Dimitrova, L. N., Kuroha, K., Tatematsu, T. & Inada, T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 (2009)
Letzring, D. P., Dean, K. M. & Grayhack, E. J. Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16, 2516–2528 (2010)
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011)
Park, S. H. et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145 (2013)
Hipp, M. S., Park, S. H. & Hartl, F. U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014)
Ong, S. E. & Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nature Protocols 1, 2650–2660 (2006)
Yan, W. & Craig, E. A. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 19, 7751–7758 (1999)
Turner, G. C. & Varshavsky, A. Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120 (2000)
Duttler, S., Pechmann, S. & Frydman, J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 (2013)
Wang, M., Herrmann, C. J., Simonovic, M., Szklarczyk, D. & von Mering, C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015)
Heck, J. W., Cheung, S. K. & Hampton, R. Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl Acad. Sci. USA 107, 1106–1111 (2010)
Summers, D. W., Wolfe, K. J., Ren, H. Y. & Cyr, D. M. The type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS ONE 8, e52099 (2013)
Amiel, J., Trochet, D., Clement-Ziza, M., Munnich, A. & Lyonnet, S. Polyalanine expansions in human. Hum. Mol. Genet. 13, R235–R243 (2004)
Forood, B., Perez-Paya, E., Houghten, R. A. & Blondelle, S. E. Formation of an extremely stable polyalanine beta-sheet macromolecule. Biochem. Biophys. Res. Commun. 211, 7–13 (1995)
Fandrich, M. & Dobson, C. M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21, 5682–5690 (2002)
Gray, M. J. et al. Polyphosphate is a primordial chaperone. Mol. Cell 53, 689–699 (2014)
Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013)
Zu, T. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013)
Roth, D. M. et al. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 12, e1001998 (2014)
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D. & Hegemann, J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23 (2002)
Young, C. L., Raden, D. L., Caplan, J. L., Czymmek, K. J. & Robinson, A. S. Cassette series designed for live-cell imaging of proteins and high-resolution techniques in yeast. Yeast 29, 119–136 (2012)
Mumberg, D., Muller, R. & Funk, M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768 (1994)
Ito-Harashima, S., Kuroha, K., Tatematsu, T. & Inada, T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 (2007)
Ito, Y., Suzuki, M. & Husimi, Y. A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem. Biophys. Res. Commun. 264, 556–560 (1999)
Scheibel, T. et al. The charged region of Hsp90 modulates the function of the N-terminal domain. Proc. Natl Acad. Sci. USA 96, 1297–1302 (1999)
Kragt, A., Voorn-Brouwer, T., van den Berg, M. & Distel, B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 280, 7867–7874 (2005)
Kryndushkin, D. S., Alexandrov, I. M., Ter-Avanesyan, M. D. & Kushnirov, V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278, 49636–49643 (2003)
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996)
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003)
Park, S. H. et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18, 153–165 (2007)
Scazzari, M., Amm, I. & Wolf, D. H. Quality control of a cytoplasmic protein complex: chaperone motors and the ubiquitin-proteasome system govern the fate of orphan fatty acid synthase subunit Fas2 of yeast. J. Biol. Chem. 290, 4677–4687 (2015)
Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003)
Acknowledgements
We thank D. Cyr, T. Inada, A. van Hoof and D. H. Wolf for reagents, and M. S. Hipp for discussions. Assistance by A. Ries and M.-J. Yoon is gratefully acknowledged. This work was supported by the European Commission under FP7 GA n°ERC-2012-SyG_318987–ToPAG, the Munich Cluster for Systems Neurology (SyNergy) and the Center for integrated Protein Science Munich (CiPSM).
Author information
Authors and Affiliations
Contributions
Y.-J.C. and S.-H.P. designed and performed most of the biochemical and functional experiments. T.H. performed the sucrose gradient and semi-denaturing detergent agarose gel electrophoresis analyses. R.K. performed the mass spectrometry analysis and proteomics and L.V.-D the bioinformatics. F.U.H. and M.H.-H. supervised the experimental design and wrote the manuscript with contributions from Y.-J.C. and the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Properties of NS-proteins in ltn1Δ cells.
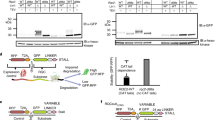
a, Firefly luciferase (Luc) or NS–Luc was expressed under the GAL1 promoter in WT or ltn1Δ yeast cells for ~16 h (~5 generations) at 30 °C. Proteins were immunoprecipitated (IP) from cell extracts with anti-Myc antibody, followed by anti-Luc immunoblotting (IB). SDS-res., SDS-resistant aggregates. The cell extracts used as input were analysed by immunoblotting against phosphoglycerate kinase 1 (Pgk1p) as a loading control. EV, empty vector. b, SDS-resistant HMW forms of NS–Luc do not represent polyubiquitylated protein. Myc-tagged NS–Luc was expressed under the GAL1 promoter in pdr5Δ or ltn1Δ yeast cells. pdr5Δ cells were incubated with DMSO or with MG132 (95 μM in DMSO) for 1.5 h. Cell lysates were prepared under denaturing conditions (see Methods), followed by NS–Luc IP with anti-myc antibody and IB with anti-Luc antibody (left panel) or anti-Ub antibody (right panel). The positions of SDS-resistant NS–Luc, polyUb–Luc and IgG are indicated. NS–Luc and Pgk1p in input fractions were analysed. c, The WT yeast strain used in this study (BY4741) and its LTN1 deletion strain were in the [RNQ+] state. To cure [RNQ+], cells were grown on YPD plates containing 3 mM guanidinium chloride (GdmCl) and subsequently streaked on YPD plates without GdmCl to isolate single colonies. The [RNQ+] prion state was confirmed by Rnq1–GFP inclusion body formation upon expression of Rnq1–GFP from CUP1 promoter by induction for 4 h with 50 μM CuSO4 during exponential growth. Live cells were analysed by fluorescence microscopy. Scale bar, 5 μm. d, NS–GFP was expressed under the GAL1 promoter in ltn1Δ cells in the [RNQ+] or [rnq−] state. Cell extracts were analysed by IP and IB with anti-GFP antibody. e, Sucrose density gradient fractionation of ltn1Δ cells expressing NS–GFP for 16–18 h. Absorbance at 254 nm indicates the position of ribosomes and polysomes (top panel). Gradient fractions were immunoblotted for the 60S protein Rpl3p with anti-Rpl3p antibody (middle panel) or anti-GFP antibody (bottom panel). SDS-resistant material was incompletely recovered, presumably due to the use of 10% TCA to precipitate the fractions before IB. Note that the immunoblot was overexposed to visualize the fractionation of SDS-resistant NS–GFP.
Extended Data Figure 2 Inclusion formation by stalled poly-basic proteins in ltn1Δ cells.
a, The disordered region from Hsp82p (residues 210 to 263) was used as an alternative spacer sequence (s*) in the stalling construct GFP–s*-K20, using GFP–s* as control. Representative live cell fluorescence images are shown and cells with visible inclusions were quantified as in Fig. 1b. GFP–s-K20 (Fig. 2a) and GFP–s*-K20 showed a similar frequency of inclusion formation. b, Live cell fluorescence microscopy of ltn1Δ cells expressing the GFP–s-polyR–mCh proteins indicated (see Fig. 2b). Cells were analysed for GFP and mCherry fluorescence. Cells with visible inclusions were quantified as in Fig. 1b. Scale bar, 5μm. c, The GFP–s-polyR–mCh proteins shown on the left were expressed in ltn1Δ cells. Cell extracts were analysed by SDD-AGE, followed by IB with anti-GFP antibody. Note that constructs R4RARE (3) and R20RARE (5) form SDS-resistant aggregates detectable by SDS–PAGE (Fig. 2b), while R20FREQR4RARE (6) forms inclusions but little SDS-resistant aggregates by SDS–PAGE (Fig. 2b). In lane 1, only 25% of cell lysate was applied to avoid overloading.
Extended Data Figure 3 Rqc2p-dependent aggregation of stalled polypeptides.
a, NS–GFP is released from ribosomes in rqc2Δ cells. Sucrose density gradient fractionation of rqc2Δ cells expressing NS–GFP for 16–18 h. Analysis was performed as in Extended Data Fig. 1e. b, Live cell fluorescence microscopy of RQC mutant cells expressing NS–GFP. Hoechst 33342 was used for nuclear staining. Cells with visible inclusions were quantified as in Fig. 1b. Scale bar, 5 μm. c, rqc2Δ cells preserve the ability to deposit aggregated protein in inclusions. The rqc2Δ and ltn1Δrqc2Δ strains used in this study were derived from the [RNQ+] WT strain. Rnq1–GFP was expressed as in Extended Data Fig. 1c to confirm inclusion formation in the RQC2 deletion strain. WT [rnq−] cells were isolated from WT [RNQ+] cells by GdmCl treatment as in Extended Data Fig. 1c. Inclusion formation was analysed by fluorescence microscopy. Scale bar, 5 μm. d, RQC2 or rqc2aaa was expressed under the RQC2 promoter in ltn1Δrqc2Δ cells expressing GFP–s-R20RARE–mCh. Cell extracts were analysed by IB with anti-GFP antibody. Pgk1p was used as a loading control. CAT-tails are indicated. e, RQC2 deletion prevents inclusion formation of stalled polypeptides in ltn1Δ cells. GFP–s-K20 was expressed in ltn1Δrqc2Δ cells under the GAL1 promoter. WT Rqc2p or Rqc2aaa was co-expressed under the RQC2 promoter in a single copy plasmid. Inclusion formation was analysed by fluorescence microscopy and quantified as in Fig. 1b. Scale bar, 5 μm.
Extended Data Figure 4 Engineered CAT-tails mediate aggregation.
a, Schematic representation of GFP–s fusion proteins with stop codon containing 20 Lys residues or a (Ala-Thr)6 sequence, or 20 Lys residues followed by a (Ala-Thr)6 or (Gly-Ser)6 sequence (top). Live cell fluorescence microscopy of ltn1Δhel2Δ cells expressing the proteins indicated. The fraction of cells with visible inclusions is indicated (quantified as in Fig. 1b). Scale bar, 5 μm. b, Deletion of HEL2 increases read-through efficiency through a 20 Lys tract (encoded by AAG codons). The fusion proteins indicated and shown schematically in the top panel were expressed in ltn1Δ or ltn1Δhel2Δ cells. Cell extracts were analysed by IB with anti-GFP antibody (bottom panel). Arrowhead indicates position of full-length GFP–s-K20–mCh.
Extended Data Figure 5 Chaperone sequestration by aggregates of stalled polypeptides.
a, SDS-resistant co-aggregates of NS–GFP with Sis1p are solubilized with formic acid. NS–GFP was expressed in ltn1Δ cells and immunoprecipitated from cell extracts with anti-GFP antibody. The precipitate was incubated without or with formic acid (FA) as in Fig. 1a and analysed by IB with anti-Sis1p antibody. b, Formation of SDS-resistant NS–GFP aggregates and co-aggregation with Sis1p are independent of prion state. NS–GFP was expressed under the GAL1 promoter in ltn1Δ [RNQ+], ltn1Δ [rnq−] or ltn1Δrqc2Δ [RNQ+] cells. Cell extracts were analysed by IP with anti-GFP, followed by IB with anti-GFP antibody (left panel) or anti-Sis1p antibody (right panel). Interaction of Sis1p and NS–GFP in [rnq−] state indicates that their interaction was not mediated by Rnq1p aggregates. c, Sis1–mCh co-localizes with NS–GFP inclusions in ltn1Δ cells. NS–GFP was expressed under the GAL1 promoter at 30 °C in cells with SIS1-mCh integrated into the SIS1 locus in the chromosome. Live cells were analysed by fluorescence microscopy. Scale bar, 5 μm. d, NS–GFP aggregation and co-aggregation with Sis1p in ltn1Δrqc2Δ cells is restored by expression of WT RQC2 but not rqc2aaa. NS–GFP was co-expressed with WT Rqc2p or Rqc2aaa in ltn1Δrqc2Δ cells. NS–GFP was expressed under the GAL1 promoter and WT Rqc2p and Rqc2aaa were expressed under the RQC2 promoter. Cell lysates were analysed by IP with anti-GFP, followed by IB with anti-GFP antibody (left panel) or anti-Sis1p antibody (right panel). e, Formation of Sis1p positive inclusions in ltn1Δ cells not expressing recombinant NS-protein. WT or ltn1Δ cells expressing Sis1–GFP from the genomic SIS1 locus were grown in YPD media at 30 °C. Cells with ≥ 2 Sis1–GFP inclusions were quantified by analysing >200 cells per condition in 4 independent experiments. Scale bar, 5 μm. f, Sis1p in ltnlΔ cells in high molecular weight (HMW) aggregates. Cell extracts from WT, ltnlΔ and ltn1Δrqc2Δ cells not expressing recombinant NS-protein were analysed by Blue native-PAGE and IB with anti-Sis1p antibody. Pgk1p was used as a loading control. The positions of the native Sis1p dimer and of HMW forms are indicated. The amount of HMW Sis1p was quantified by densitometry and expressed as percent of total. Error bars, s.d. from three independent experiments. P values from Student’s t-test. g, Aggregation of Sis1p in ltn1Δrqc2Δ cells is restored by expression of WT RQC2 but not rqc2aaa. Extracts of ltn1Δrqc2Δ cells expressing WT Rqc2p or Rqc2aaa under the RQC2 promoter were analysed as in f without expression of recombinant NS-protein. h, Formation of SDS-resistant aggregates in ltn1Δ cells observed with Sis1–HA or Sis1–GFP. SIS1 was chromosomally replaced by SIS1-HA or SIS1-GFP in WT or ltn1Δ cells. Tagged Sis1 proteins were immunoprecipitated with anti-HA antibody or anti-GFP antibody, followed by IB with anti-Sis1p antibody (right panel). Input fraction was analysed with anti-Sis1p antibody (left panel). Gel slices corresponding to the position of SDS-resistant Sis1p aggregates were excised from gels and subjected to MS-analysis to identify proteins interacting with the aggregates (see Methods). i, Proteins in SDS-resistant Sis1p aggregates are of relatively high abundance in the total yeast proteome. Abundance values measured in total proteome in ppm are plotted33.
Extended Data Figure 6 Impairment of cytosolic protein quality control in ltn1Δ cells.
a, CmCh* ubiquitylation is preserved in ltn1Δ cells. Following expression of CmCh* from GAL1 promoter, His6-tagged ubiquitin (His6–Ub) expression from CUP1 promoter was induced with CuSO4 for 4 h before harvesting cells. Ubiquitylated proteins were isolated by His6 pull-down from cell lysates prepared in 6 M GdmCl to preserve polyubiquitylation. Eluates were analysed by IB with anti-CPY antibody. The positions of CmCh* and of polyubiquitylated CmCh* (CmCh*–Ubn) are indicated. b, Inhibition of degradation of CPY* in ltn1Δ cells is rescued by overexpression of Sis1p. CPY* fused to GFP (CG*) and N-terminally HA-tagged Sis1p (HA–Sis1p) were expressed from the GAL1 promoter. The degradation of CG* was analysed after inhibition of protein synthesis with cycloheximide as in Fig. 5a. CG* was detected by IB with anti-GFP antibody and Sis1p with anti-HA antibody (top panel). Pgk1p was used as a loading control. Data were quantified by densitometry (bottom panel). Error bars, s.d. from three independent experiments. c, Sis1p co-aggregates with CmCh* in ltn1Δ cells. CPY*–mCherry (CmCh*) was expressed under the GAL1 promoter at 30 °C in ltn1Δ cells expressing Sis1–GFP from the genomic SIS1 locus. Live cells were analysed by fluorescence microscopy. Scale bar, 5 μm. d, Live cell fluorescence microscopy of WT and ltn1Δ cells co-expressing NS–GFP with CmCh* for 18 h at 30 °C. Nuclei were counterstained with Hoechst 33342. DIC, differential interference contrast. Scale bar, 5 μm. e, Sis1p overexpression does not suppress the formation of SDS-resistant NS–GFP aggregates in ltn1Δ cells. NS–GFP and N-terminally HA-tagged Sis1p (HA–Sis1p) was expressed under the GAL1 promoter in WT or ltn1Δ cells. Empty vector was used as a control for HA–Sis1p. SDS-resistant aggregates were analysed by IP with anti-GFP, followed by IB with anti-GFP antibody (left panel) or anti-Sis1p antibody (right panel).
Extended Data Figure 7 Additional proteostasis stress causes growth defect of ltn1Δ cells.
a–c, Growth phenotype of RQC mutant strains. Cells from WT and RQC mutant strains indicated were transformed with CmCh* expression vector under the GAL1 promoter and were serially fivefold diluted before spotting onto glucose medium (−Induction) and galactose/raffinose medium (+Induction). Plates were incubated for 3 days at 37 °C. In b, galactose inducible HA-tagged Sis1p was expressed. EV, empty vector. d, Hygromycin B (HygB) sensitivity of RQC mutant strains. Cells from WT and RQC mutant strains indicated were grown to exponential phase in liquid YPD medium, serially fivefold diluted and spotted onto YPD plates with or without HygB (18 μg ml−1). Plates without HygB were incubated for 2 days and with HygB for 3 days at 37 °C. e, Formation of Sis1p positive inclusions in ltn1Δ cells is enhanced under proteotoxic stress. ltn1Δ cells expressing Sis1–GFP replacing chromosomal SIS1 were grown in YPD media at 30 °C without or with hygromycin B (400 μg ml−1) for 18 h. Live cell fluorescence microscopy was performed. Cells with Sis1–GFP inclusions were quantified by analysing >200 cells per condition in 4 independent experiments. Data shown are an extension of the experiment shown in Extended Data Fig. 5e. Scale bar, 5 μm.
Supplementary information
Supplementary Information
This file contains full legends for Supplementary Tables 1 and 2 (see separate files for tables) and Supplementary Figure 1, which contains full-size gel images. (PDF 1071 kb)
Supplementary Data
This file contains Supplementary table 1 – see Supplementary Information document for full legend. (XLSX 126 kb)
Supplementary Data
This file contains Supplementary Table 2 – see Supplementary Information document for full legend. (XLSX 123 kb)
Rights and permissions
About this article
Cite this article
Choe, YJ., Park, SH., Hassemer, T. et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016). https://doi.org/10.1038/nature16973
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16973
This article is cited by
-
Valosin containing protein (VCP): initiator, modifier, and potential drug target for neurodegenerative diseases
Molecular Neurodegeneration (2023)
-
Proteomics revealed an association between ribosome-associated proteins and amyloid beta deposition in Alzheimer's disease
Metabolic Brain Disease (2023)
-
Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis
Nature (2022)
-
Lsm7 phase-separated condensates trigger stress granule formation
Nature Communications (2022)
-
The role of the glycerol transporter channel Fps1p in cellular proteostasis during enhanced proteotoxic stress
Applied Microbiology and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.