Abstract
Haematopoietic stem cells (HSCs), which sustain production of all blood cell lineages1, rely on glycolysis for ATP production2,3, yet little attention has been paid to the role of mitochondria. Here we show in mice that the short isoform of a critical regulator of HSCs, Prdm16 (refs 4, 5), induces mitofusin 2 (Mfn2), a protein involved in mitochondrial fusion and in tethering of mitochondria to the endoplasmic reticulum. Overexpression and deletion studies, including single-cell transplantation assays, revealed that Mfn2 is specifically required for the maintenance of HSCs with extensive lymphoid potential, but not, or less so, for the maintenance of myeloid-dominant HSCs. Mfn2 increased buffering of intracellular Ca2+, an effect mediated through its endoplasmic reticulum–mitochondria tethering activity6,7, thereby negatively regulating nuclear translocation and transcriptional activity of nuclear factor of activated T cells (Nfat). Nfat inhibition rescued the effects of Mfn2 deletion in HSCs, demonstrating that negative regulation of Nfat is the prime downstream mechanism of Mfn2 in the maintenance of HSCs with extensive lymphoid potential. Mitochondria therefore have an important role in HSCs. These findings provide a mechanism underlying clonal heterogeneity among HSCs8,9,10,11 and may lead to the design of approaches to bias HSC differentiation into desired lineages after transplantation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008)
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010)
Takubo, K. et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61 (2013)
Aguilo, F. et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood 117, 5057–5066 (2011)
Chuikov, S., Levi, B. P., Smith, M. L. & Morrison, S. J. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nature Cell Biol. 12, 999–1006 (2010)
de Brito, O. M. & Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 (2008)
Rizzuto, R., De Stefani, D., Raffaello, A. & Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nature Rev. Mol. Cell Biol. 13, 566–578 (2012)
Müller-Sieburg, C. E., Cho, R. H., Thoman, M., Adkins, B. & Sieburg, H. B. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood 100, 1302–1309 (2002)
Sanjuan-Pla, A. et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502, 232–236 (2013)
Muller-Sieburg, C. E. & Sieburg, H. B. Clonal diversity of the stem cell compartment. Curr. Opin. Hematol. 13, 243–248 (2006)
Dykstra, B. et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 1, 218–229 (2007)
Chan, D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 (2012)
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012)
Tanaka, A. & Youle, R. J. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol. Cell 29, 409–410 (2008)
Shing, D. C. et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 117, 3696–3707 (2007)
Yoshida, M. et al. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood 103, 2753–2760 (2004)
Sorianello, E. et al. The promoter activity of human Mfn2 depends on Sp1 in vascular smooth muscle cells. Cardiovasc. Res. 94, 38–47 (2012)
Beerman, I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl Acad. Sci. USA 107, 5465–5470 (2010)
Weksberg, D. C., Chambers, S. M., Boles, N. C. & Goodell, M. A. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood 111, 2444–2451 (2008)
Pham, A. H., McCaffery, J. M. & Chan, D. C. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843 (2012)
Chen, H., McCaffery, J. M. & Chan, D. C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130, 548–562 (2007)
Szilvassy, S. J., Humphries, R. K., Lansdorp, P. M., Eaves, A. C. & Eaves, C. J. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc. Natl Acad. Sci. USA 87, 8736–8740 (1990)
Ngoh, G. A., Papanicolaou, K. N. & Walsh, K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J. Biol. Chem. 287, 20321–20332 (2012)
Li, H., Rao, A. & Hogan, P. G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103 (2011)
Müller, M. R. et al. Requirement for balanced Ca/NFAT signaling in hematopoietic and embryonic development. Proc. Natl Acad. Sci. USA 106, 7034–7039 (2009)
Wu, H., Peisley, A., Graef, I. A. & Crabtree, G. R. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 17, 251–260 (2007)
Aramburu, J. et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285, 2129–2133 (1999)
Benz, C. et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10, 273–283 (2012)
Dykstra, B., Olthof, S., Schreuder, J., Ritsema, M. & de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208, 2691–2703 (2011)
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003)
Zambrowicz, B. P. et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl Acad. Sci. USA 100, 14109–14114 (2003)
Pham, A. H., McCaffery, J. M. & Chan, D. C. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843 (2012)
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA 93, 5860–5865 (1996)
Chen, H., McCaffery, J. M. & Chan, D. C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130, 548–562 (2007)
Shimshek, D. R. et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26 (2002)
Ausubel, F. M. Current Protocols in Molecular Biology (Wiley, 1987)
Benz, C. et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10, 273–283 (2012)
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003)
Sorianello, E. et al. The promoter activity of human Mfn2 depends on Sp1 in vascular smooth muscle cells. Cardiovasc. Res. 94, 38–47 (2012)
Grynkiewicz, G., Poenie, M. & Tsien, R. Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 (1985)
Acknowledgements
The pHR-IRES-GFP SIN vector was a gift from R. Creusot. Mfn2−/− MEFs were a gift from D. Chan. We thank S.-H. Ho and E. A. Schon for assistance and discussion. This work was supported by grant NIH RO1 CA167286 and RO1 AG029262 (H.-W.S.), the Druckenmiller Fellowship from the New York Stem Cell Foundation (L.L.L.), the Ruth L. Kirschstein F31 CA196045 fellowship (D.J.C.), the NIH National Center for Research Resources award 1S10RR027050-01 and the NIH Office of the Director award 1S10OD020056-01.
Author information
Authors and Affiliations
Contributions
L.L.L. designed and performed most experiments, contributed to the concept, and co-wrote the manuscript with H.-W.S.; M.J.A. generated the data in Fig. 4e and Extended Data Fig. 8a, and assisted in Figs 2, 4i and Extended Data Figs 6, 7, 8. M.M. cloned the DNDrp1 construct and verified its function (Extended Data Fig. 7d). D.J.C. generated the Mfn2 promoter construct (Extended Data Fig. 3c). H.-W.S. provided concept and guidance, and co-wrote the paper with L.L.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
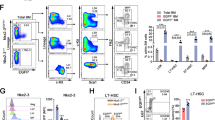
Extended Data Figure 1 Representative sort gates for isolation of HSCs.
Flow cytometric plots showing the gates used to isolate HSCs (Lin−Sca1+kit+Flt3−CD48−) and CD150hi and CD150lo HSCs.
Extended Data Figure 2 Mitochondrial dynamics in Prdm16−/− MEFs.
a, Frequency distribution (left) and frequency distribution profile (right) of mitochondrial length in WT or Prdm16−/− fetal HSCs. Bars, mean ± s.e.m.; n ≥ 20 fields of cells from three biological replicates; *P < 0.05 within length bins; two-tailed Student’s t-test within length bins. b, Mitochondrial morphology in Prdm16−/− MEFs treated for 24 h with the Drp1 inhibitor, mDivi (30 μM), or vehicle is shown for comparison, Mitotracker Red staining, scale bar 20 μm). c, Frequency distribution (left) and frequency distribution profile (right) of mitochondrial length in WT and Prdm16−/− MEFs treated for 24 h with the Drp1 inhibitor, mDivi (30 μM), or vehicle. Bars, mean ± s.e.m.; n ≥ 16 fields from three biological replicates; *P < 0.05 within length bins; one-way ANOVA with Bonferroni’s post-hoc test within length bins. d, Fluorescence micrographs showing fusion hybrids of WT or Prdm16−/− MEFs transduced with baculovirus expressing mitochondria-tagged GFP and RFP before PEG-mediated fusion (scale bar 50 μm). Fused mitochondria are yellow (arrows), and were only observed in fusions of WT cells. e, western blot (upper, Supplementary Fig. 1 for full scans) and quantification of western blots for Mfn2 of WT and Prdm16−/− MEFs. Bars, mean ± s.e.m.; n = 3 fields biological replicates; *P < 0.05; two-tailed Student’s t-test. f, Mitochondrial morphology in WT, Prdm16−/− and Mfn2−/− MEFs visualized by Tomm20 staining (red) (scale bar 20 μm). (G) Mitochondrial length in WT and Prdm16−/− MEFs transduced with EV or Mfn2-IRES-GFP (Mfn2–GFP). Bars, mean ± s.e.m.; n ≥ 24 fields from three biological replicates; *P < 0.05 compared to Prdm16−/− EV in each length bin; one-way ANOVA with Bonferroni’s post-hoc test within length bins.
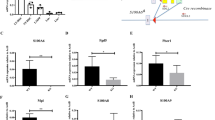
Extended Data Figure 3 sPrdm16 interacts with the Mfn2 promoter.
a, Schematic representation of Prdm16 protein domain structure. b, qPCR probe amplification scheme covering the Mfn2 promoter used in chromatin immunoprecipitation assays. c, Proximal Mfn2 promoter luciferase gene reporter assay in WT MEFs transfected 24 h previously with flPrdm16, sPrdm16 or control vectors. Bars, mean ± s.e.m.; n = 3 biological replicates; *P < 0.05; one-way ANOVA with Dunnett’s post-hoc test. d, ChIP quantification in Prdm16−/− MEFs transduced with retroviral Flag–Prdm16 constructs immunoprecipitated using Flag and TF2D antibodies. TFIID positive (GAPDH) and negative (intergenic Chr 6) control probes are also shown. Quantification was performed after establishment of qPCR standard curves for all probes (see Methods). Bars, mean ± s.e.m.; n = 3 technical replicates representative tow biological replicates; *P < 0.05; one-way ANOVA with Dunnett’s post-hoc test. e, Percentage CD45.2 (donor) contribution in PB WBC in CD45.1+CD45.2+ mice reconstituted with 200 transduced (IRES–GFP or Mfn2-IRES–GFP) Prdm16+/− CD45.2+ HSCs and 2 × 105 CD45.1+ competitor bone marrow cells. Plots, mean ± s.e.m.; n = 17 recipients pooled from four independent transplants; *P < 0.05; two-tailed Student’s t-test.
Extended Data Figure 4 Mitochondrial morphology of bone marrow populations.
a, Representative images of mitochondrial morphology in Pham-reporter+ CD150hi and CD150lo HSCs (lin−Sca1+kit+Flt3−CD48−), MPPs (lin−Sca1+kit+CD48+), CMPs (lin−Sca1−kit+), CLPs (lin−Sca1lokitloIL7Ra+Flt3+) and lineage+ cells (scale bar, 5 μm). b, Mitochondrial length frequency distribution in Pham-reporter+ HSCs, progenitors and Lin+ cells. Bars, mean ± s.e.m.; n ≥ 14 fields from three biological replicates; *P < 0.05 within length bins; one-way ANOVA with Bonferroni’s post-hoc test within length bins.
Extended Data Figure 5 Phenotype and function of Mfn2−/− HSCs.
a, Mitochondrial morphology HSCs from Mfn2fl/fl or Mfn2fl/fl-Vav-Cre (Mfn2−/−) mice visualized with immunostaining for Tomm20. b, Frequency distribution of mitochondrial lengths (right panel, compared to WT in each length bin). Bars, mean ± s.e.m.; n ≥ 7 fields from two biological replicates; *P < 0.05 within length bins; two-tailed Student’s t-test within length bins. c, CD150 surface staining mean fluorescence intensity (MFI) of HSCs (Lin−Sca1+kit+CD48−) in 8–12 week old Mfn2fl/fl or Mfn2fl/fl.Vav-Cre (Mfn2−/−) mice. n = 25 biological replicates, *P < 0.05, two-tailed Student’s t-test. d, e, CD150 MFI of Mfn2fl/fl or Mfn2fl/fl.Vav-Cre (Mfn2−/−) donor HSCs 15 weeks after competitive transplantation. Plots in d, mean ± s.e.m.; n ≥ 15 mice from three biological replicates. f, *P < 0.05; two-tailed Student’s t-test. f, Donor (CD45.2) chimaerism 15 weeks after competitive transplantation of 2 × 105 Mfn2fl/fl or Mfn2fl/fl.Vav-Cre (Mfn2−/−) fetal liver cells together with 2 × 105 CD45.1+ competitor bone marrow cells into CD45.1+CD45.2+ recipients Plots, mean ± s.e.m.; n = 10 mice pooled from two independent transplants; *P < 0.05; two-tailed Student’s t-test.
Extended Data Figure 6 Analysis of competitive repopulation experiments and lentiviral vector transduction.
a, Schematic representation of lentiviral constructs used. b, Representative example of transduction efficiency of purified HSCs with IRES–GFP and IRES–mCherry vectors 24–48 h post-transduction. c, Schematic representation of transduction and subsequent competitive repopulation experiments. d, Representative flow cytometric plots of the gates used to analyse recipients of competitive repopulation experiments. The CD45.1/CD45.2 gates were also applied to the myeloid, B and T cell gates to determine donor contribution to individual lineages. In this example, HSCs had been transduced with an IRES–GFP lentiviral vectors (efficiency 80%, see b). Although donor repopulation was high in the periphery (upper left panel, 71.8%) no GFP was detected (lower left panel), suggesting silencing of the vectors. e, Quantification of proviral copy number in donor-derived (CD45.2+) 15 weeks after transduction and competitive transplantation of HSCs. Despite silencing of the vector (see d), approximately 1 proviral copy was present per donor HSC-derived cell. n = 3 biological replicates; *P < 0.05; two-tailed Student’s t-test. f, Representative example of transduction efficiency of purified HSCs with IRES–GFP and Mfn2–IRES–GFP lentiviral vectors. g, Expression of Mfn2 mRNA in CD150hi and CD150lo HSCs 24 h post-transduction relative to EV control (left panel) and in donor-derived HSCs, CMPs and lineage+ cells 15 weeks after competitive transplantation of 200 transduced HSCs together with 2 × 105 CD45.1+ competitor cells (right panel). The data indicate partial silencing of the vector in HSCs, and complete silencing in the progenitors and more mature cells. n = 3 biological replicates; *P < 0.05; two-way analysis of variance (ANOVA) with Bonferroni’s post-hoc test. h, Donor (CD45.2+) chimaerism 15 weeks after transplantation of 200 CD45.2+ HSCs transduced for 24 h with SIN LTR constructs pLVX-IRES-GFP or pLVX-Mfn2-IRES-GFP together with 2 × 105 CD45.1+ competitor fetal liver cells into lethally irradiated CD45.1+CD45.2+ recipients mice. Plots, mean ± s.e.m.; n ≥ 19 recipients from four transplantation experiments; *P < 0.05; two-tailed Student’s t-test. i, Same experiments as in h, but using the non-SIN LTR vector, pHR. Plots, mean ± s.e.m.; n = 5 recipients; *P < 0.05; two-tailed Student’s t-test.
Extended Data Figure 7 Nfat activity in Mfn2−/− and Prdm16−/− MEFs.
a, Mfn2fl/fl (blue) or Mfn2fl/fl-Vav-Cre (Mfn2−/−) (red) HSCs were stained with DCFDA to visualize intracellular reactive oxygen species levels. Cells were treated with oligomycin for 15 min as a positive control. b, HSC apoptosis levels were analysed by surface staining of Annexin V. c, Intracellular staining for the endoplasmic reticulum stress response chaperone GRP78 in Mfn2fl/fl or Mfn2fl/fl-Vav-Cre (Mfn2−/−) HSCs (red colours) MPPs (green colours) and CMPs (blue colours). Histograms; representative of three biological replicates. d, Calcium flux trace (left) and baseline intracellular Ca2+ (right) in WT LSK cells transduced with IRES–GFP or Mfn2–IRES–GFP lentiviral vectors (e) and with WT or Mfn2−/− MEFs stained with Indo1. Bars, mean ± s.e.m.; n = 3 biological replicates; *P < 0.05; two-tailed Student’s t-test. f, Nfat1 staining (left, scale bars, 5 μm) and fraction of nuclear Nfat1 (right, *P < 0.05) in HSCs transduced with IRES–GFP or Mfn2–IRES–GFP lentiviral vector. Bars, mean ± s.e.m.; n ≥ 10 from two biological replicates; *P < 0.05; two-tailed Student’s t-test. g, Nfat1 staining (left; scale bars, 20 μm) and fraction of nuclear Nfat1 (right; *P < 0.05) in WT or Mfn2−/− MEFs. Bars, mean ± s.d.; n ≥ 20 fields from three independent experiments; *P < 0.05; two-tailed Student’s t-test. h, Nfat luciferase gene reporter activity in WT or Mfn2−/− MEFs. Bars, mean ± s.d.; n = 3 experiments; *P < 0.05; two-tailed Student’s t-test. i, Mitochondrial morphology after transfection of dominant negative Drp1 into HeLa cells (scale bars, 5 μm). Note extreme elongation of mitochondria (arrow), confirming that this vector is functional. j, Nfat1 staining (left, scale bars, 5 μm) and fraction of nuclear Nfat1 (right) in HSCs lentivirally transduced with sPRrdm16 and flPRdm16. Plots, mean ± s.e.m.; n ≥ 14 fields of cells pooled from two biological replicates; *P < 0.05; one-way ANOVA with Dunnett’s post-hoc test. k, Nfat luciferase gene reporter activity in WT and in Prdm16−/− MEFs transfected with Mfn2, flPrdm16 or sPrdm16 constructs. Bars, mean ± s.d.; n = 3 experiments; *P < 0.05; one-way ANOVA with Dunnett’s post-hoc test.
Extended Data Figure 8 Effect of VIVIT on WT HSCs.
a, Expression of lymphoid (Sox4 and Il7r) and myeloid/platelet (Vwf and Fli1) genes in HSCs treated with VIVIT or DMSO for 24 h. n = 3 biological replicates; *P < 0.05; Student’s t-test. b, Lymphoid/myeloid ratio of CD45.2+ HSCs cultured for 4 days in DMSO and 500 nM VIVIT, transplanted with 2 × 105 CD45.1+CD45.2+ competitor bone marrow cells into CD45.1+ recipients and analysed 15 weeks post-transplant for lymphoid/myeloid ratio of donor compartment. Plots, mean ± s.e.m.; n = 5–9 recipients from two transplant experiments; *P < 0.05; two-tailed Student’s t-test. c, Donor chimaerism analysis 15 weeks after transplantation of CD45.2+ HSCs transduced with IRES–GFP or VIVIT–GFP with 2 × 105 CD45.1+CD45.2+ competitor bone marrow cells into CD45.1+ recipients Plots, mean ± s.e.m.; n = 9–12 recipients from two independent transplant experiments; *P < 0.05; two-tailed Student’s t-test. Bottom panel shows lymphoid and myeloid donor contribution in each individual recipient.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Tables 1- 3. (PDF 331 kb)
Source data
Rights and permissions
About this article
Cite this article
Luchsinger, L., de Almeida, M., Corrigan, D. et al. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 529, 528–531 (2016). https://doi.org/10.1038/nature16500
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16500
This article is cited by
-
MITOL deficiency triggers hematopoietic stem cell apoptosis via ER stress response
The EMBO Journal (2024)
-
Emerging roles of mitochondrial functions and epigenetic changes in the modulation of stem cell fate
Cellular and Molecular Life Sciences (2024)
-
Mitochondrial fusion is a therapeutic vulnerability of acute myeloid leukemia
Leukemia (2023)
-
Transcription factor Nkx2-3 maintains the self-renewal of hematopoietic stem cells by regulating mitophagy
Leukemia (2023)
-
Small molecule agonist of mitochondrial fusion repairs mitochondrial dysfunction
Nature Chemical Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.