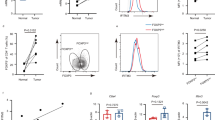
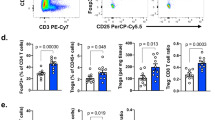
Abstract
Regulatory T (Treg) cells expressing the transcription factor Foxp3 have a pivotal role in maintaining immunological self-tolerance1,2,3,4,5; yet, excessive Treg cell activities suppress anti-tumour immune responses6,7,8. Compared to the resting Treg (rTreg) cell phenotype in secondary lymphoid organs, Treg cells in non-lymphoid tissues exhibit an activated Treg (aTreg) cell phenotype9,10,11. However, the function of aTreg cells and whether their generation can be manipulated are largely unexplored. Here we show that the transcription factor Foxo1, previously demonstrated to promote Treg cell suppression of lymphoproliferative diseases12,13, has an unexpected function in inhibiting aTreg-cell-mediated immune tolerance in mice. We find that aTreg cells turned over at a slower rate than rTreg cells, but were not locally maintained in tissues. aTreg cell differentiation was associated with repression of Foxo1-dependent gene transcription, concomitant with reduced Foxo1 expression, cytoplasmic localization and enhanced phosphorylation at the Akt sites. Treg-cell-specific expression of an Akt-insensitive Foxo1 mutant prevented downregulation of lymphoid organ homing molecules, and impeded Treg cell homing to non-lymphoid organs, causing CD8+ T-cell-mediated autoimmune diseases. Compared to Treg cells from healthy tissues, tumour-infiltrating Treg cells downregulated Foxo1 target genes more substantially. Expression of the Foxo1 mutant at a lower dose was sufficient to deplete tumour-associated Treg cells, activate effector CD8+ T cells, and inhibit tumour growth without inflicting autoimmunity. Thus, Foxo1 inactivation is essential for the migration of aTreg cells that have a crucial function in suppressing CD8+ T-cell responses; and the Foxo signalling pathway in Treg cells can be titrated to break tumour immune tolerance preferentially.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008)
Shevach, E. M. Biological functions of regulatory T cells. Adv. Immunol. 112, 137–176 (2011)
Josefowicz, S. Z., Lu, L. F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012)
Benoist, C. & Mathis, D. Treg cells, life history, and diversity. Cold Spring Harb. Perspect. Biol. 4, a007021 (2012)
Bluestone, J. A., Bour-Jordan, H., Cheng, M. & Anderson, M. T cells in the control of organ-specific autoimmunity. J. Clin. Invest. 125, 2250–2260 (2015)
Zou, W. Regulatory T cells, tumour immunity and immunotherapy. Nature Rev. Immunol. 6, 295–307 (2006)
Nishikawa, H. & Sakaguchi, S. Regulatory T cells in tumor immunity. Int. J. Cancer 127, 759–767 (2010)
Quezada, S. A., Peggs, K. S., Simpson, T. R. & Allison, J. P. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol. Rev. 241, 104–118 (2011)
Huehn, J. et al. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199, 303–313 (2004)
Miyara, M. et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009)
Sugiyama, D. et al. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl Acad. Sci. USA 110, 17945–17950 (2013)
Ouyang, W. et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012)
Kerdiles, Y. M. et al. Foxo transcription factors control regulatory T cell development and function. Immunity 33, 890–904 (2010)
Smigiel, K. S. et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 211, 121–136 (2014)
Mueller, S. N., Gebhardt, T., Carbone, F. R. & Heath, W. R. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31, 137–161 (2013)
Hedrick, S. M., Hess Michelini, R., Doedens, A. L., Goldrath, A. W. & Stone, E. L. FOXO transcription factors throughout T cell biology. Nature Rev. Immunol. 12, 649–661 (2012)
Ouyang, W. & Li, M. O. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. 32, 26–33 (2011)
Luo, C. T. & Li, M. O. Transcriptional control of regulatory T cell development and function. Trends Immunol. 34, 531–539 (2013)
Fabre, S. et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J. Immunol. 181, 2980–2989 (2008)
Kerdiles, Y. M. et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nature Immunol. 10, 176–184 (2009)
Ouyang, W., Beckett, O., Flavell, R. A. & Li, M. O. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009)
Curiel, T. J. et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nature Med. 10, 942–949 (2004)
Shimizu, J., Yamazaki, S. & Sakaguchi, S. Induction of tumor immunity by removing CD25+CD4+ T cells: a common basis between tumor immunity and autoimmunity. J. Immunol. 163, 5211–5218 (1999)
Bos, P. D., Plitas, G., Rudra, D., Lee, S. Y. & Rudensky, A. Y. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J. Exp. Med. 210, 2435–2466 (2013)
Franklin, R. A. et al. The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014)
Stringer, J. R. et al. Modeling variation in tumors in vivo. Proc. Natl Acad. Sci. USA 102, 2408–2413 (2005)
Bauer, C. A. et al. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J. Clin. Invest. 124, 2425–2440 (2014)
Levine, A. G., Arvey, A., Jin, W. & Rudensky, A. Y. Continuous requirement for the TCR in regulatory T cell function. Nature Immunol. 15, 1070–1078 (2014)
Vahl, J. C. et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014)
Ali, K. et al. Inactivation of PI(3)K p110δ breaks regulatory T-cell-mediated immune tolerance to cancer. Nature 510, 407–411 (2014)
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008)
Wan, Y. Y., Chi, H., Xie, M., Schneider, M. D. & Flavell, R. A. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nature Immunol. 7, 851–858 (2006)
Wright, D. E., Wagers, A. J., Gulati, A. P., Johnson, F. L. & Weissman, I. L. Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936 (2001)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Stewart, T. J. & Abrams, S. I. Altered immune function during long-term host-tumor interactions can be modulated to retard autochthonous neoplastic growth. J. Immunol. 179, 2851–2859 (2007)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012)
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014)
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013)
Harrow, J. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7 Suppl 1, 1–9 (2006)
Acknowledgements
We thank A. Rudensky for the Foxp3Cre mouse strain and S. Abrams for the AT-3 cell line. We also thank the Li laboratory for discussions. This work was supported by the National Institutes of Health (RO1 AI102888-01A1 to M.O.L.), Ludwig Center for Cancer Immunology (M.O.L.), and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Author information
Authors and Affiliations
Contributions
C.T.L. and M.O.L. were involved in all aspects of this study including planning and performing experiments, analysing and interpreting data, and writing the manuscript. W.L. processed and analysed next-generation sequencing data, and wrote the manuscript. S.D. and A.T. participated in mouse colony management and experiments including parabiosis. M.O.L. oversaw all the work performed.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Parabiotic analysis of Treg cells from different organs.
a, Flow cytometric analysis of CD44 and CD62L in Treg cells (CD4+Foxp3+) from lymph node, spleen, liver and colon lamina propria of C57BL/6 mice. Percentages of rTreg and aTreg cells are shown. A sizable population of intermediate phenotype (CD62LhiCD44hi) Treg cells was also present in the liver. b, Graphical representation of the parabiosis experiments. Congenically mismatched C57BL/6 mice were surgically connected (time point −4). Parabionts were analysed 2 or 4 weeks after surgery (time point −2 or 0). In separation experiments, parabiotic mice that had been connected for 4 weeks were surgically disconnected from each other (time point 0). Separated mice were analysed 2, 6, 12 or 18 weeks after surgery (time point 2, 6, 12 or 18). c, Representative flow cytometric plots showing chimaerism of naive CD4+ T cells or different Treg cell subsets in a CD45.2/2+ parabiont at various time points.
Extended Data Figure 2 aTreg cell differentiation is associated with Akt-triggered suppression of Foxo1.
a, Gene expression comparison of Foxo1-regulated genes in splenic aTreg versus rTreg cells. Foxo1-regulated genes were defined by the following criteria: (1) differentially expressed between wild-type and Foxo1-knockout Treg cells; (2) the expression was corrected by expression of a constitutively active mutant of Foxo1 (Foxo1CA). b, Normalized expression of all transcripts isolated from splenic aTreg cells were plotted against transcripts from splenic rTreg cells. Some of the Foxo1 direct target genes were highlighted. c, Comparison of Lamc1, Nid2, Mmp9 mRNA levels in rTreg versus aTreg subsets. d, Flow cytometric analysis of phosphorylated S6 ribosomal protein in splenic aTreg and rTreg cells. Quantification shows mean fluorescence intensity normalized to rTreg cells. n = 3. *P < 0.05 (paired t-test). e, aTreg cell differentiation experiments in vitro. rTreg cells (CD4+Foxp3+CD62LhiCD44lo) from spleen and lymph nodes of Foxp3-YFP reporter mice were purified by flow cytometric sorting, and were stimulated with anti-CD3/CD28 and IL-2 for 3 days. Akt inhibitor (Akti, MK-2206) or DMSO solvent control were added to the culture. Phosphorylated Foxo1, and CD44 and CD62L levels were determined by flow cytometry. Quantification shows percentages of rTreg and aTreg cells in DMSO control or Akt inhibitor group. n = 3. *P < 0.05 (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 3 Foxo1 hyperactivation in Treg cells does not affect expression of activation markers or proliferation.
a, b, Treg cells (CD4+Foxp3+) from spleens and lymph nodes of wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice were purified by flow cytometric sorting. a, Haemagglutinin (HA)-tagged Foxo1CA protein in the whole-cell lysate was measured by immunoblotting with anti-HA. b, Subcellular localization of HA–Foxo1CA was determined by immunofluorescence staining with anti-HA. c, Flow cytometric analysis of Foxo1-activated target genes CCR7 and Bim in lymph node Treg cells from 9–12-day-old wild-type, CA/+ and CA/CA mice. Grey shaded lines represent isotype controls. d, Flow cytometric analysis of ICOS and CD62L expression in lymph node Treg cells from 9–12-day-old wild-type, CA/+ and CA/CA mice. Bar graph shows fractions of ICOS+CD62Lhi, ICOS+CD62Llo subsets among Treg cells. n = 4. e, CD62LhiCD44lo rTreg cells from wild-type or CA/CA mice, or CD62LloCD44hi aTreg cells from wild-type mice were isolated by flow cytometric sorting, and the expression of Cd69, Egr2 and Il1r2 mRNA was determined by reverse transcriptase quantitative PCR (RT–qPCR). f, EdU was administrated intraperitoneally into 9–10-day-old wild-type or CA/CA mice. EdU incorporation in lymph node rTreg and aTreg cells was analysed 18 h after injection. n = 3. **P < 0.01 (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 4 CA-expressing Treg cells show intact activation, proliferation and survival in vitro.
CD4+Foxp3+CD62LhiCD44loCD69−ICOS− rTreg cells were isolated from wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice, and were stimulated with anti-CD3, anti-CD28 and IL-2 for 3 days. a, CD44, CD62L, CD69 and ICOS expression was determined by flow cytometry. Grey shaded lines represent isotype controls. b, Cells were stained with CellTrace Violet dye before culture, and cell division was tracked by dilution of the dye. Grey shaded line represents undivided cells. c, Cell death was measured by propidium iodide (PI) incorporation. Percentages of live cell fraction (PI−) were compared. n = 4. NS, not significant (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 5 Foxo1 hyperactivation depletes aTreg cells in mice.
a, In vivo labelling experiment: anti-CD4-biotin (RM4-4 clone) was injected intravenously into wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice, and spleens were collected 5 min after injection. A non-competing clone of anti-CD4 (RM4-5) was used in subsequent flow cytometric analysis to label total CD4+ T cells. Representative flow cytometric plots from gated conventional CD4+ T cells (CD4 RM4-5+Foxp3−) are shown. Quantification shows percentage of RM4-4+ among conventional CD4+ T cells. n = 10 (WT, CA/CA); n = 7 (CA/+). b, c, The frequencies of lymph node rTreg or aTreg cells among CD4+ T cells of 9–12-day-old wild-type, CA/+ or CA/CA mice. n = 4–6. c, The numbers of lymph node rTreg or aTreg cells of 9–12-day-old wild-type, CA/+ or CA/CA mice. n = 6 (WT, CA/CA); n = 4 (CA/+). Unpaired t-test. Data are mean ± s.e.m.
Extended Data Figure 6 Foxo1 hyperactivation preferentially impairs aTreg cells, but does not affect Treg cell suppressive function in vitro.
a, The frequencies of lymph node and splenic rTreg or aTreg cells among CD4+ T cells of 4–6-week-old wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice. n = 10. b, The numbers of lymph node rTreg or aTreg cells of 4–6-week-old wild-type, CA/+ or CA/CA mice. n = 10. c, A representative picture of 5-week-old wild-type, CA/+ or CA/CA mice. d, Body weight of 4–6-week-old wild-type, CA/+ and CA/CA mice. n = 5. e, The numbers of CD4+ and CD8+ T cells from lymph node and spleen of 4–6-week-old wild-type, CA/+ and CA/CA mice. n = 7 (WT, CA/+); n = 14 (CA/CA). f, Flow cytometric analysis of CTLA4, LAG3 and GITR expression in lymph node rTreg and aTreg cells from 4–6-week-old wild-type, CA/+ and CA/CA mice. Grey shaded lines represent isotype controls. g, Suppression of wild-type naive CD4+ T cells, labelled with CellTrace Violet (Tresp, responding T cells), by wild-type, CA/+ or CA/CA Treg cells. Tresp cell division was assessed by CellTrace Violet dilution at the indicated ratios of cell numbers between Treg and Tresp cells. Grey line represents conditions without Treg cell in culture. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 7 Modest increase of inflammatory cytokine production by CD4+ and CD8+ T cells in Foxp3CreFoxo1CA/Foxo1CA mice.
a, d, Flow cytometric analysis of CD44 and CD62L expression in CD4+Foxp3− conventional T cells (a) or CD8+ T cells (d) from lymph node and spleen of 4–6-week-old wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice. b, c, Representative histogram (b), and quantification (c) of cytokine (IFN-γ, IL-4, IL-17) expression by CD4+Foxp3− conventional T cells from lymph node and spleen of 4–6-week-old wild-type, CA/+ and CA/CA mice. n = 14 (WT, CA/CA for IFN-γ), n = 7 (WT, CA/CA for IL-4, IL-17); n = 3 (CA/+). e, f, Representative histogram (e), and quantification (f) of IFN-γ expression by CD8+ T cells from lymph node and spleen of 4–6-week-old wild-type, CA/+ and CA/CA mice. n = 9 (WT); n = 6 (CA/+); n = 11 (CA/CA). *P < 0.05; **P < 0.01 (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 8 Characterization of CD8+ T cells from non-lymphoid tissues.
a, b, Flow cytometric analysis (a), and quantification (b) of IFN-γ expression by CD8+ T cells from liver and colon lamina propria of 4–6-week-old wild-type, Foxp3CreFoxo1CA/+ (CA/+) or Foxp3CreFoxo1CA/Foxo1CA (CA/CA) mice. n = 6 (WT); n = 3 (CA/+); n = 8 (CA/CA). c, CD8+ to CD4+ T-cell ratio among lymph node, spleen, liver and lamina propria T cells of 4–6-week-old wild-type, CA/+ and CA/CA mice. n = 10 (WT); n = 8 (CA/+); n = 13 (CA/CA). *P < 0.05 (unpaired t-test). Data are mean ± s.e.m.
Extended Data Figure 9 CD8+ T-cell depletion rescues the lethal disease in Foxp3CreFoxo1CA/Foxo1CA mice.
a, A representative image of 5-week-old wild-type, CD8−/−, Foxp3CreFoxo1CA/Foxo1CA and Foxp3CreFoxo1CA/Foxo1CACD8−/− mice. b, Haematoxylin and eosin staining of liver and colon sections from 5-week-old wild-type, CD8−/−, Foxp3CreFoxo1CA/Foxo1CA and Foxp3CreFoxo1CA/Foxo1CACD8−/− mice.
Extended Data Figure 10 Foxp3CreFoxo1CA/+ mice show enhanced anti-tumour immune responses.
a, Flow cytometric analysis of CD44 and CD62L in Treg cells (CD4+Foxp3+) from lymph node and tumour of PyMT tumour-bearing mice. b–d, Eight-to-ten-week-old wild-type or Foxp3CreFoxo1CA/+ (CA/+) mice received orthotopic inoculation of PyMT-derived mammary tumour cells (AT-3). b, Survival of wild-type and CA/+ mice received tumour implantation. c, Foxp3 expression in tumour-infiltrating CD4+ T cells from day 30–35 AT-3 tumour. d, Flow cytometric analysis of GzmB expression in CD8+ T cells from wild-type or CA/+ tumour-bearing mice. e, Tumour growth curve of wild-type, CD8−/−, CA/+, or Foxp3CreFoxo1CA/+ CD8−/− (CA/+CD8−/−) mice that received orthotopic inoculation of AT-3 tumour cells. n = 4. *P < 0.05; **P < 0.01; ****P < 0.0001; log-rank test (b) and two-way analysis of variance (ANOVA) (e). Data are mean ± s.e.m.
Supplementary information
Supplementary Table
This file contains Supplementary Table 1. (XLSX 159 kb)
Rights and permissions
About this article
Cite this article
Luo, C., Liao, W., Dadi, S. et al. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature 529, 532–536 (2016). https://doi.org/10.1038/nature16486
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16486
This article is cited by
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Proinflammatory plasticity towards Th17 paradigm of regulatory T cells consistent with elevated prevalence of TGFBR2 variants in elderly patients with primary immune thrombocytopenia
BMC Immunology (2023)
-
Comprehensive analysis of the lncRNAs-related immune gene signatures and their correlation with immunotherapy in lung adenocarcinoma
British Journal of Cancer (2023)
-
Intricacies of TGF-β signaling in Treg and Th17 cell biology
Cellular & Molecular Immunology (2023)
-
RIPK1 plays a crucial role in maintaining regulatory T-Cell homeostasis by inhibiting both RIPK3- and FADD-mediated cell death
Cellular & Molecular Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.