Abstract
During ageing, muscle stem-cell regenerative function declines. At advanced geriatric age, this decline is maximal owing to transition from a normal quiescence into an irreversible senescence state. How satellite cells maintain quiescence and avoid senescence until advanced age remains unknown. Here we report that basal autophagy is essential to maintain the stem-cell quiescent state in mice. Failure of autophagy in physiologically aged satellite cells or genetic impairment of autophagy in young cells causes entry into senescence by loss of proteostasis, increased mitochondrial dysfunction and oxidative stress, resulting in a decline in the function and number of satellite cells. Re-establishment of autophagy reverses senescence and restores regenerative functions in geriatric satellite cells. As autophagy also declines in human geriatric satellite cells, our findings reveal autophagy to be a decisive stem-cell-fate regulator, with implications for fostering muscle regeneration in sarcopenia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cheung, T. H. & Rando, T. A. Molecular regulation of stem cell quiescence. Nature Rev. Mol. Cell Biol. 14, 329–340 (2013)
Comai, G. & Tajbakhsh, S. Molecular and cellular regulation of skeletal myogenesis. Curr. Top. Dev. Biol. 110, 1–73 (2014)
Yin, H., Price, F. & Rudnicki, M. A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013)
Montarras, D., L’Honore, A. & Buckingham, M. Lying low but ready for action: the quiescent muscle satellite cell. FEBS J. 280, 4036–4050 (2013)
Grounds, M. D. Therapies for sarcopenia and regeneration of old skeletal muscles: more a case of old tissue architecture than old stem cells. Bioarchitecture . 4, 81–87 (2014)
García-Prat, L., Sousa-Victor, P. & Muñoz-Cánoves, P. Functional dysregulation of stem cells during aging: a focus on skeletal muscle stem cells. FEBS J. 280, 4051–4062 (2013)
Chakkalakal, J. V., Jones, K. M., Basson, M. A. & Brack, A. S. The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 (2012)
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014)
Sousa-Victor, P., Garcia-Prat, L., Serrano, A. L., Perdiguero, E. & Muñoz-Cánoves, P. Muscle stem cell aging: regulation and rejuvenation. Trends Endocrinol. Metab. 26, 287–296 (2015)
Cosgrove, B. D. et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nature Med. 20, 255–264 (2014)
Bernet, J. D. et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nature Med. 20, 265–271 (2014)
Price, F. D. et al. Inhibition of JAK–STAT signaling stimulates adult satellite cell function. Nature Med. 20, 1174–1181 (2014)
Tierney, M. T. et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nature Med. 20, 1182–1186 (2014)
Cuervo, A. M. et al. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy 1, 131–140 (2005)
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009)
Fukada, S. et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459 (2007)
Liu, L. et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep . 4, 189–204 (2013)
Pallafacchina, G. et al. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res . 4, 77–91 (2010)
Carnio, S. et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep . 8, 1509–1521 (2014)
Rubinsztein, D. C., Marino, G. & Kroemer, G. Autophagy and aging. Cell 146, 682–695 (2011)
Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. & Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 (2004)
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008)
Zhu, J., Dagda, R. K. & Chu, C. T. Monitoring mitophagy in neuronal cell cultures. Methods Mol. Biol. 793, 325–339 (2011)
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo . Cell Metab. 6, 458–471 (2007)
Morselli, E. et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615–629 (2011)
van Deursen, J. M. The role of senescent cells in ageing. Nature 509, 439–446 (2014)
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007)
Muñoz-Espín, D. & Serrano, M. Cellular senescence: from physiology to pathology. Nature Rev. Mol. Cell Biol. 15, 482–496 (2014)
Blagosklonny, M. V. Selective anti-cancer agents as anti-aging drugs. Cancer Biol. Ther. 14, 1092–1097 (2013)
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009)
Narita, M. et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011)
Pérez-Mancera, P. A., Young, A. R. & Narita, M. Inside and out: the activities of senescence in cancer. Nature Rev. Cancer 14, 547–558 (2014)
Capparelli, C. et al. Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis via glycolysis and ketone production. Cell Cycle 11, 2285–2302 (2012)
Flach, J. et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014)
Kodama, R. et al. ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic Ras-induced premature senescence. Genes Cells 18, 32–41 (2013)
Ramsey, M. R. & Sharpless, N. E. ROS as a tumour suppressor? Nature Cell Biol. 8, 1213–1215 (2006)
Ito, K. et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature Med. 12, 446–451 (2006)
Lee, A. C. et al. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J. Biol. Chem. 274, 7936–7940 (1999)
Mandal, P. K., Blanpain, C. & Rossi, D. J. DNA damage response in adult stem cells: pathways and consequences. Nature Rev. Mol. Cell Biol. 12, 198–202 (2011)
Shao, L. et al. Reactive oxygen species and hematopoietic stem cell senescence. Int. J. Hematol. 94, 24–32 (2011)
Lerner, C. et al. Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts. Aging Cell 12, 966–977 (2013)
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013)
Warr, M. R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013)
Tang, A. H. & Rando, T. A. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 33, 2782–2797 (2014)
Lee, J. H. et al . Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228 (2010)
Masiero, E. et al. Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515 (2009)
Suelves, M. et al. uPA deficiency exacerbates muscular dystrophy in MDX mice. J. Cell Biol. 178, 1039–1051 (2007)
Breitkreutz, B. J., Jorgensen, P., Breitkreutz, A. & Tyers, M. AFM 4.0: a toolbox for DNA microarray analysis. Genome Biol. 2, http://dx.doi.org/10.1186/gb-2001-2-8-software0001 (2001)
Hulsen, T., de Vlieg, J. & Alkema, W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 (2008)
Sacco, A. et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143, 1059–1071 (2010)
Crews, L. et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of α-synucleinopathy. PLoS ONE 5, e9313 (2010)
Perdiguero, E. et al. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38α in abrogating myoblast proliferation. EMBO J. 26, 1245–1256 (2007)
Kamentsky, L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011)
Bolte, S. & Cordelieres, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006)
Acknowledgements
We are indebted to G. Mariño for the gift of GFP–LC3 transgenic mice, C. Keller and M. Capecchi for Pax–Cre mouse lines, J. Ruberte for TEM studies help, E. Masliah and K. Kosberg for Atg7 lentivirus; M. Raya, M. Jardí, and V. Lukesova for their technical contributions, and especially J. Guerra for help in microarray experiments and P. Sousa-Victor for initial findings; J. Martín-Caballero (PRBB Animal Facility) and O. Fornas (UPF/CRG FACS Facility) for technical help, and the KS Society. The authors acknowledge funding from MINECO, Spain (SAF2012-38547, SAF2015-67369-R, PLE2009-0124; SAF2009-08374; “María de Maeztu” Programme for Units of Excellence in R&D MDM-2014-0370), AFM, E-Rare/ERANET, Fundació Marató TV3, MDA, EU-FP7 (Myoage, Optistem and Endostem) and DuchennePP-NL. M.M.-V. acknowledges funding from ISCIII, Spain (FIS-PS09/01267, FIS-PI13/02512, CP09/00184, PI14/01529) and CIBERNED; and MS from the European Union ERC (282310-MyoPHAGY) and Foundation Leducq. L.G.-P. was supported by a Predoctoral Fellowship from Programa de Formación de Personal Investigador (Spain).
Author information
Authors and Affiliations
Contributions
L.G.-P. designed and performed most experiments, analysed data, interpreted results and wrote the manuscript. A.L.S. and E.P. designed and performed experiments, and helped in interpreting results and editing the manuscript. M.M.-V. and M.S. helped in designing and interpreting some experiments and results and editing the manuscript. L.O., V.R.-B. and S.G. performed some experiments and provided technical support. E.R. provided technical support in microscopy. J.R.-U. and E.B. performed ChIP experiments and helped in interpreting results. P.M.-C. conceived the project, designed experiments, interpreted results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
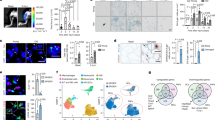
Extended Data Figure 1 The reduced autophagy flux in quiescent satellite cells can be increased by pharmacological treatment in vivo.
a, Venn diagrams of overlapping genes between a proteostasis gene set (See Supplementary Table 1) and genes significantly upregulated in quiescent satellite cells from the indicated publications or from our gene expression microarray data comparing freshly FACS isolated satellite cells from resting muscle, or muscles obtained 72 h after cardiotoxin (CTX) injury, from young, wild-type mice. b, K-means clustering analysis (performed with Gene-E, Broad Institute) of the gene expression of the autophagy-related genes during ageing. Clusters are shown with heat maps of the normalized raw data. Each column represents a different sample and each row a different gene probe. Red, increased expression; white, neutral expression; blue, decreased expression. c, Representative example of the FACS strategy and gating scheme for isolating satellite cells from mice in resting conditions. d, Pax7 and GFP immunostaining of freshly isolated satellite cells from resting muscles of young and old GFP–LC3 mice. Scale bar, 5 μm. e, Electron microscopy images of young and old satellite cells on sections of resting tibialis anterior (TA) muscle of wild-type (WT) mice. Arrowheads indicate autophagic vesicles. Scale bars, 1 μm and 0.5 μm (right and left, respectively). f, Pax7 and GFP immunostaining on tissue sections from resting tibialis anterior muscles of young and old GFP–LC3 mice. Arrowheads indicate autophagic vesicles. Scale bar, 5 μm. g, p62 and ubiquitin (Ub) MFI. Arrowheads, co-localization of p62 and ubiquitin aggregates. h, LC3 western blot of freshly isolated satellite cells from young and old, wild-type mice, treated with bafilomycin or vehicle for 4 h before collection. Graph shows LC3II quantification, after normalization with tubulin levels; for full scan see Supplementary Fig. 2. i, Quiescent satellite cells were freshly isolated from old, wild-type mice subjected to two weeks of rapamycin, spermidine or vehicle (control) treatment. Cells were treated (or not treated) with bafilomycin 4 h prior to analysis by immunostaining of LC3 marker. Z projections of representative fluorescence microscopy images are shown. Scale bars, 5 μm. j, Representative fluorescent microscopy images from Fig. 1d. Scale bar, 5 μm. Data show mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-test. P values are indicated. Number of samples were n = 3 animals per group for a and b; n = 35 (young) and 66 (old) cells analysed from 3 animals for g; n = 3 animals per group for h.
Extended Data Figure 2 Reinduction of autophagy rescues proliferation and reduces senescence in geriatric satellite, thus restoring regenerative capacity.
a, Transplanted muscles from Fig. 2c were immunostained for GFP and for Ki67, Pax7, MyoD or Mgn (to determine the distinct possible myogenic states of satellite cells in the regenerating muscle). Scale bars, 50 μm. b, Autophagy flux analysed by flow cytometry in freshly isolated satellite cells from resting muscle of GFP–LC3 mice, treated for 48 h with rapamycin or vehicle (control). Satellite cells were treated with bafilomycin or vehicle for 4 h before analysis. Results are expressed as the change in GFP–LC3 MFI in bafilomycin (−) compared to bafilomycin (+) conditions. c, Western blot analysis of pS6 protein levels in geriatric satellite cells from wild-type mice, treated for 48 h with rapamycin or vehicle (control). Graph shows pS6 quantification, normalized to tubulin; for full scan see Supplementary Information Fig. 2. d, As in Fig. 2c, percentage γH2AX+ or p16INK4a+cells from total GFP+ cells were quantified. Scale bars, 10 μm. e, Quantification BrdU+ and senescence-associated β-gal+ satellite cells, pre-treated as in Fig. 2c and analysed after 96 h. f, Quantification of senescent (senescence-associated β-gal+) satellite cells, isolated from young and geriatric wild-type mice, pre-treated for 48 h with spermidine or vehicle (control) and cultured for 96 h. g, Quantitative real-time PCR (RT–qPCR) analysis of Atg7 expression on satellite cells infected with LV-Atg7 or LV-control (LV-Co), and cultured for 96 h. h, GFP–LC3 satellite cells were infected with LV-Atg7 or LV-Co and treated with bafilomycin or vehicle for 4 h before analysis. Autophagy flux was analysed by flow cytometry and represented as in b. Representative images are shown. Scale bar, 10 μm. i, Muscle regeneration experiment by satellite cell transplantation. An equal number of satellite cells from young and geriatric mice infected with a lentivirus overexpressing the Atg7 gene (LV-Atg7) or a lentivirus control (LV-Co), which also expressed GFP, were transplanted into injured muscle of young immunodeficient mice, and collected 28 days later. GFP expression in muscles was analysed by immunostaining. Quantification of GFP+ cells (fibres) per muscle field versus transplanted control-treated satellite cells. Representative images are shown. Scale bar, 75 μm. j, EDL geriatric muscles, infected with LV-Atg7 or LV-Co, and grafted on recipient mouse muscle. Regeneration was analysed 8 days later. Frequency distribution of regenerating fibres by size. Scale bar, 25 μm. Data are mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-test. P values are indicated. Number of samples were n = 60,000 cells analysed from 3 animals for b; n = 3 animals per group for c; n = 5 engraftments per group for d; n = 3 animals per group for e–g; n = 60,000 cells analysed from 3 animals for h; n = 3 engraftments per group for i; n = 4 engraftments per group for j.
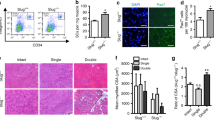
Extended Data Figure 3 Genetic impairment of autophagy in young quiescent satellite cells leads to premature senescence and impaired muscle regeneration.
a, RT–qPCR analysis of Atg7 expression and western blot analysis of LC3, p62 and tubulin of satellite cells isolated from Atg7WT and Atg7ΔPax7 mice. Graph shows the quantification of p62 normalized to tubulin; for full scan see Supplementary Fig. 2. b, Quiescent satellite cells were freshly isolated from Atg7WT and Atg7ΔPax7 mice which had been subjected to two weeks of rapamycin or vehicle (control) treatment in vivo. Cells were treated (or controls were untreated) with bafilomycin 4 h before analysis by fluorescence microscopy. Z projections of representative fluorescence microscopy images are shown. Scale bar, 5 μm. c, Quantification of satellite cells in resting muscle of three-month-old Atg7WT and Atg7ΔPax7 mice by flow cytometry analysis (α7 integrin+CD34+ cells per gram of muscle tissue). d, Representative fluorescent microscopy images from Fig. 3d. Scale bar, 10 μm. e, RT–qPCR analysis of MyoD, Mgn and Ki67 expression in freshly isolated quiescent satellite cells from resting muscle of Atg7WT and Atg7ΔPax7ER mice, 7 days after tamoxifen treatment. f, Percentage of activated satellite cells (Pax7+/MyoD+) from the total Pax7+ cells (FACS-isolated 14-h post-injury from (a)). Scale bar, 50 μm. g, pS6 and Lamp1 immunostaining of cells from a. Scale bar, 10 μm. h, γH2AX protein levels per nucleus in Pax7+ satellite cells in tibialis anterior muscles of Atg7WT and Atg7ΔPax7ER mice, 15 days post-injury. Representative images are shown. Scale bar, 25 μm. i, Pax7+ satellite cells were quantified following immunostaining on regenerating muscles of Atg7WT and Atg7ΔPax7ER mice 7 days and 15 days after cardiotoxin injury. j, Representative images of haematoxylin and eosin staining of muscles at 7 days post-injury on muscles of Atg7WT and Atg7ΔPax7ER mice. Fibre size of central-nucleated myofibres at 7 days and 28 days post-injury is quantified. Scale bar, 50 μm. k, Tibialis anterior muscles of Atg7WT and Atg7ΔPax7 mice were injured by cardiotoxin injection and 21 days later these muscles were reinjured and then subsequently analysed 21 days later (21 + 21 days post-injury). The size of central-nucleated myofibres was quantified. Representative images are shown. Scale bar, 50 μm. l, Pax7+ and Ki67+ double-positive satellite cells were quantified following immunostaining on regenerating muscles of Atg7WT and Atg7ΔPax7ER mice 7 days after cardiotoxin injury. m, An equal number of quiescent satellite cells from Atg7WT:GFP–LC3 and Atg7ΔPax7:GFP–LC3 mice (two weeks ± rapamycin pre-treatment), transplanted as in Fig. 2c, and immunostained with the indicated antibodies 4 days later. Quantification of GFP+ cells per muscle field. Values relative to transplanted young cells (100%). Representative images are shown. Scale bar, 75 μm. n, Percentage of GFP+ cells that are also Ki67+ cells in muscles from m. o, Quantification of proliferating (BrdU+) and senescent (SA-β-gal+) satellite cells, isolated from Atg7WT and Atg7ΔPax7, pre-treated for 48 h with spermidine or rapamycin (or control vehicle) and cultured for 96 h. Data show mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-test. P values are indicated. The number of samples were n = 3 animals per group (a); n = 7 animals per group (c); n = 3 animals per group (e–l); n = 4 engraftments per group (m, n); n = 3 animals per group (o).
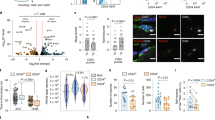
Extended Data Figure 4 Autophagy loss in satellite cells causes dysfunctional mitophagy and mitochondria accumulation, leading to increased ROS and senescence.
a, p62 and ubiquitin immunostaining on freshly isolated satellite cells from resting muscle of three-month-old Atg7WT and Atg7ΔPax7ER mice, one month after tamoxifen treatment. Arrowheads indicate co-localization of p62 and Ub aggregates. Representative images are shown. Scale bar, 5 μm. b, TOM20 and Lamp1 immunostaining of quiescent satellite cells isolated from young and geriatric WT mice. Mice were subjected to two weeks of rapamycin, spermidine or Trolox (or vehicle) treatment before analysis. Co-localization was calculated as the area occupied by the immunofluorescence co-localizing staining on images with respect to the total cellular area. The Pearson’s coefficient (r) was used to analyse the correlation of the intensity values of green and red pixels in the dual-channel images. The z projections of representative fluorescence microscopy images are shown. Scale bar, 5 μm. c, Mitochondria quantification by MitoTracker in quiescent satellite cells of old mice, treated with rapamycin or vehicle for two weeks. d, Mitochondria (MitoTracker labelling) in young or geriatric cells. Satellite cells, were pre-treated with CCCP for 1 h (see Methods) and ± rapamycin for 24 h. Percentage of MitoTracker MFI reduction ± rapamycin. e, For the mitochondrial membrane potential analysis, satellite cells were freshly isolated from young wild-type mice and treated for 1 h with CCCP or DMSO (control). Membrane potential (TMRM MFI/MitoTracker Green MFI ratio) of cells was calculated by flow cytometry analysis at 1 h and 24 h after CCCP treatment (being 100% the membrane potential value of control satellite cells). f, Mitochondria content was quantified by MitoTracker staining of satellite cells from young and geriatric wild-type mice treated with rapamycin or vehicle (control) for 48 h. The z projections of representative fluorescence microscopy images are shown. Scale bar, 5 μm. g, Mitochondria and ROS detection by MitoTracker and CellROX staining, respectively. Co-localization was calculated as in b. The z projections of representative fluorescence microscopy images are shown. Scale bar, 5 μm. h, Representative images of freshly isolated satellite cells from resting muscle of three-month-old Atg7WT and Atg7ΔPax7 mice stained with CellROX fluorescent dye and p16INK4a antibody. Scale bar, 5 μm. Data are mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-test. P values are indicated. Number of samples were n = 36 (Atg7WT) and n = 38 (Atg7ΔPax7ER) cells analysed from 3 animals (a); n = 23 (young), n = 24 (control), n = 42 (rapamycin); n = 28 (spermidine) and n = 21 (Trolox) cells analysed from 3 animals (b); n = 60,000 cells analysed from 3 animals (c); n = 40,000 cells analysed from 4 animals (d); n = 30,000 cells analysed from 3 animals (e, f); n = 18 (young), 21 (control), 15 (rapamycin) and 13 (Trolox) cells analysed from 3 animals (g).
Extended Data Figure 5 ROS inhibition in autophagy-impaired aged and Atg7 null satellite cells significantly restores cell proteostasis.
a, ROS-level quantification in quiescent satellite cells from three-month-old Atg7WT and Atg7ΔPax7 mice by CellROX flow cytometry. Representative images are shown. Scale bar, 5 μm. b, Western blot analysis of 53BP1 and parkin in satellite cells isolated from three-month-old Atg7WT and Atg7ΔPax7 mice. Tubulin control is the same tubulin control for Fig. 3g. Graph shows quantification of 53BP1 and parkin protein normalized to tubulin; for full scan see Supplementary Information Fig. 1. c, Quantification of ROS levels for satellite cells isolated from young and geriatric WT mice by flow cytometry using CellROX fluorescent dye. Satellite cells were treated with Trolox or vehicle (control) for 48 h before analysis. Results are represented as variation of MFI between young and geriatric satellite cells. d, Quantification of p62 and ubiquitin protein levels on immunostained freshly isolated satellite cells from resting muscle of old wild-type mice, in vivo treated for 2 weeks with Trolox or vehicle (control). Representative images are shown. Scale bar, 5 μm. e, Western blot analysis of LC3 and tubulin in satellite cells isolated from geriatric WT mice and treated for 48 h with Trolox or vehicle (control), in the absence or presence of bafilomycin for 4 h before analysis. Graph shows quantification of LC3II protein normalized to tubulin; for full gel scan see Supplementary Information Fig. 2. f, Autophagy flux and mitochondria in satellite cells from GFP–LC3 mice (two weeks with or without Trolox treatment). Satellite cells treated for 4 h ± bafilomycin treatment. Representative images are shown. Scale bar, 5 μm. g, The mRFP–GFP–LC3 plasmid was transfected into young or geriatric satellite cells, with 48 h treatment ± Trolox and then 4 h treatment ± bafilomycin, prior to fixation. The percentage of autophagosomes was quantified as in Fig. 2a. h, Muscle regeneration using geriatric satellite cell transplantation. An equal number of freshly isolated geriatric satellite cells, infected with GFP lentivirus and treated for 48 h with Trolox or vehicle, were transplanted into injured muscle of young immunodeficient mice. Four days later, muscles were collected and immunostained for GFP, MyoD and Mgn (to determine the possible myogenic states of satellite cells in the regenerating muscle). Representative images are shown. Scale bar, 50 μm. i, ChIP analysis for H2AK119ub (H2Aub) in satellite cells isolated from young and geriatric wild-type mice. j, Quantification of proliferating (BrdU+) and senescent (SA-β-gal+) satellite cells isolated from Atg7WT and Atg7ΔPax7 mice treated 48 h with Trolox or vehicle (control) and cultured for 96 h. k, Quantification of proliferating (BrdU+) and senescent (senescence-associated β-gal+) satellite cells isolated from Atg7WT and Atg7ΔPax7 mice and infected with LV-sh p16INK4a or LV-sh scramble, and cultured for 96 h. Data show mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-tests. P values are indicated. Number of samples were n = 60,000 cells analysed from 3 animals (a); n = 3 animals per group (b); n = 60,000 cells analysed from 3 animals (c); n = 36 (control) and n = 35 (Trolox) cells analysed from 3 animals (d); n = 3 animals per group (e); n = 60,000 cells analysed from 3 animals (f); n = 21 (young), n = 20 (young, Trolox), n = 19 (young, + bafilomycin), n = 18 (young, Trolox + bafilomycin), n = 21 (geriatric), n = 19 (geriatric, Trolox), n = 15 (geriatric, + bafilomycin) and n = 37 (geriatric, Trolox + bafilomycin) cells analysed from 3 animals (g); n = 3 animals per group (i–k).
Extended Data Figure 6 Effects of p16INK4a silencing in autophagy-impaired young murine satellite cells.
a, Western blotting quantification of Atg7ΔPax7 satellite cells, infected with lentiviral LV-sh-p16INK4a or LV-sh-scramble and analysed 96 h later; for full gel scan see Supplementary Fig. 2. b, Atg7WT and Atg7ΔPax7 EDL, infected with LV-sh-p16INK4a or LV-sh-scramble, and grafted as Extended Data Fig. 2j. Representative eMHC-immunostaining. Scale bar, 25 μm. Data show mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-test. P values are indicated. The number of samples were n = 3 animals per group (a) and n = 4 engraftments per group (b).
Extended Data Figure 7 Impaired autophagic flux in human geriatric satellite cells.
a, Representative images of haematoxylin and eosin staining of human muscle biopsies from young (25 years old) and geriatric (95 years old) donors in resting conditions. Arrowheads indicate atrophic myofibres. Scale bar, 50 μm. b, CD56 and p16INK4a immunostaining on human muscle sections of samples described in a. Scale bar, 10 μm. c, Western blotting analysis of p62 protein in human satellite cells from young (about 25 years old) and geriatric (over 75 years old) donors, treated for 48 h ± rapamycin; for full gel scan see Supplementary Fig. 2. d, ROS and mitochondrial content analysis in human cells from treated for 48 h ± rapamycin. Graphs show MFI variation. Scale bar, 5 μm. e, Representative images from CellROX staining from d. Scale bar, 5 μm. f, Quantification of SA-β-gal+ human cells treated for 48 h ± rapamycin. Quantification was carried out 96 h after treatment. Scale bar, 200 μm. g, Quantification of proliferating (BrdU+) young and geriatric human satellite cells in culture. Representative pictures are shown. Scale bar, 25 μm. h, Western blot analysis of pS6, total S6 and tubulin in young and geriatric human satellite cells treated for 48 h with rapamycin or vehicle (control). Graphs show p62 quantification normalized to tubulin; for full scan see Supplementary Information Fig. 2. i, Immunostaining of pS6 in young and geriatric human satellite cells treated as in h. Scale bar, 75 μm. j, Scheme showing the proposed model of how age-impaired autophagy leads to muscle stem-cell senescence and regenerative decline. Data show mean ± s.e.m. Comparisons by two-sided Mann–Whitney U-tests. P values are indicated. The number of samples were n = 3 human donors per group (a–c); n = 60,000 cells analysed from 3 human donors (d), n = 3 human donors per group (f–h).
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-2. Supplementary Figure 1 contains uncropped Western blot scans with size indications corresponding to Figures 3g, 5e, 6c and Extended Data Figure 5b. Supplementary Figure 2 contains uncropped Western blot scans with size indications corresponding to Extended Data Figures 6a, 7c, 1h, 2c, 3a, 5e and Figure 7i. (PDF 1496 kb)
Supplementary Table
This file contains Supplementary Table 1, a gene list of proteostasis stem cells; Gene list of quiescence genes belonging to each proteostasis pathway, including autophagy pathways and genes; Dataset of upregulated genes in quiescent muscle. (XLSX 181 kb)
3D reconstruction of a young satellite cell from GFP-LC3 mice.
Quiescent young satellite cells were isolated by FACS from 3-months old GFP-LC3 mice and fixed on glass slides for fluorescence microscopy analysis. 30 z stacks approximately were taken for video reconstruction as explained in the Methods section. (MP4 7006 kb)
3D reconstruction of an old satellite cell from GFP-LC3 mice.
Quiescent old satellite cells were isolated by FACS from 24-months old GFP-LC3 mice and fixed on glass slides for fluorescence microscopy analysis. 30 z stacks approximately were taken for video reconstruction as explained in the Methods section. (MP4 7124 kb)
3D reconstruction of an old satellite cell from GFP-LC3 mice treated with vehicle (control) and Bafilomycin 4 hours prior fixation.
Quiescent old satellite cells were treated for 4 hours with Bafilomycin and were isolated by FACS from 24-months old GFP-LC3 mice treated with vehicle (control) (i.e., the control of the Ramaycin treatment in Video 4) for two weeks in vivo. After sorting, satellite cells were fixed on glass slides for fluorescence microscopy analysis. 30 z stacks approximately were taken for video reconstruction as explained in the Methods section. (MP4 11029 kb)
3D reconstruction of an old satellite cell from GFP-LC3 mice treated with Rapamycin and Bafilomycin 4 hours prior fixation.
Quiescent old satellite cells were treated for 4 hours with Bafilomycin and were isolated by FACS from 24-months old GFP-LC3 mice treated with Rapamycin for two weeks in vivo. After sorting, satellite cells were fixed on glass slides for fluorescence microscopy analysis. 30 z stacks approximately were taken for video reconstruction as explained in the Methods section. (MP4 7378 kb)
Rights and permissions
About this article
Cite this article
García-Prat, L., Martínez-Vicente, M., Perdiguero, E. et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). https://doi.org/10.1038/nature16187
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16187
This article is cited by
-
Emerging roles of mitochondrial functions and epigenetic changes in the modulation of stem cell fate
Cellular and Molecular Life Sciences (2024)
-
MicroRNAs-associated with FOXO3 in cellular senescence and other stress responses
Biogerontology (2024)
-
Molecular mechanisms of tumor resistance to radiotherapy
Molecular Cancer (2023)
-
Compensatory cross-talk between autophagy and glycolysis regulates senescence and stemness in heterogeneous glioblastoma tumor subpopulations
Acta Neuropathologica Communications (2023)
-
Mechanisms of metabolic stress induced cell death of human oligodendrocytes: relevance for progressive multiple sclerosis
Acta Neuropathologica Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.