Abstract
Mycobacterium tuberculosis, a major global health threat, replicates in macrophages in part by inhibiting phagosome–lysosome fusion, until interferon-γ (IFNγ) activates the macrophage to traffic M. tuberculosis to the lysosome. How IFNγ elicits this effect is unknown, but many studies suggest a role for macroautophagy (herein termed autophagy), a process by which cytoplasmic contents are targeted for lysosomal degradation1. The involvement of autophagy has been defined based on studies in cultured cells where M. tuberculosis co-localizes with autophagy factors ATG5, ATG12, ATG16L1, p62, NDP52, BECN1 and LC3 (refs 2, 3, 4, 5, 6), stimulation of autophagy increases bacterial killing6,7,8, and inhibition of autophagy increases bacterial survival1,2,4,6,7. Notably, these studies reveal modest (~1.5–3-fold change) effects on M. tuberculosis replication. By contrast, mice lacking ATG5 in monocyte-derived cells and neutrophils (polymorponuclear cells, PMNs) succumb to M. tuberculosis within 30 days4,9, an extremely severe phenotype similar to mice lacking IFNγ signalling10,11. Importantly, ATG5 is the only autophagy factor that has been studied during M. tuberculosis infection in vivo and autophagy-independent functions of ATG5 have been described12,13,14,15,16,17,18. For this reason, we used a genetic approach to elucidate the role for multiple autophagy-related genes and the requirement for autophagy in resistance to M. tuberculosis infection in vivo. Here we show that, contrary to expectation, autophagic capacity does not correlate with the outcome of M. tuberculosis infection. Instead, ATG5 plays a unique role in protection against M. tuberculosis by preventing PMN-mediated immunopathology. Furthermore, while Atg5 is dispensable in alveolar macrophages during M. tuberculosis infection, loss of Atg5 in PMNs can sensitize mice to M. tuberculosis. These findings shift our understanding of the role of ATG5 during M. tuberculosis infection, reveal new outcomes of ATG5 activity, and shed light on early events in innate immunity that are required to regulate disease pathology and bacterial replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Deretic, V. Autophagy in tuberculosis. Cold Spring Harb. Persp. Med. 4, a018481 (2014)
Dutta, R. K., Kathania, M., Raje, M. & Majumdar, S. IL-6 inhibits IFN-γ induced autophagy in Mycobacterium tuberculosis H37Rv infected macrophages. Int. J. Biochem. Cell Biol. 44, 942–954 (2012)
Juárez, E. et al. NOD2 enhances the innate response of alveolar macrophages to Mycobacterium tuberculosis in humans. Eur. J. Immunol. 42, 880–889 (2012)
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012)
Seto, S., Tsujimura, K., Horii, T. & Koide, Y. Autophagy adaptor protein p62/SQSTM1 and autophagy-related gene Atg5 mediate autophagosome formation in response to Mycobacterium tuberculosis infection in dendritic cells. PLoS ONE 8, e86017 (2013)
Sakowski, E. T. et al. Ubiquilin 1 promotes IFN-γ-induced xenophagy of Mycobacterium tuberculosis . PLoS Pathog. 11, e1005076 (2015)
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004)
Wang, J. et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 9, e1003697 (2013)
Castillo, E. F. et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc. Natl Acad. Sci. USA 109, E3168–E3176 (2012)
Cooper, A. M. et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178, 2243–2247 (1993)
Nandi, B. & Behar, S. M. Regulation of neutrophils by interferon-γ limits lung inflammation during tuberculosis infection. J. Exp. Med. 208, 2251–2262 (2011)
Choi, J. et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935 (2014)
Hwang, S. et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 (2012)
Zhao, Z. et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008)
Martinez, J. et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature Cell Biol. 17, 893–906 (2015)
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad. Sci. USA 104, 14050–14055 (2007)
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biol. 8, 1124–1132 (2006)
Maskey, D. et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nature Commun. 4, 2130 (2013)
Jakubzick, C. et al. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J. Exp. Med. 205, 2839–2850 (2008)
Abram, C. L., Roberge, G. L., Hu, Y. & Lowell, C. A. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014)
Flynn, J. L. & Chan, J. Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129 (2001)
Jayaswal, S. et al. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA Screen. PLoS Pathog. 6, e1000839 (2010)
Jung, C. H. et al. ULK-Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009)
Mariño, G. et al. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 120, 2331–2344 (2010)
Lin, H. H. et al. Dynamic involvement of ATG5 in cellular stress responses. Cell Death Dis. 5, e1478 (2014)
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008)
Abdel Fattah, E., Bhattacharya, A., Herron, A., Safdar, Z. & Eissa, N. T. Critical role for IL-18 in spontaneous lung inflammation caused by autophagy deficiency. J. Immunol. 194, 5407–5416 (2015)
Kanayama, M., He, Y.-W. & Shinohara, M. L. The lung is protected from spontaneous inflammation by autophagy in myeloid cells. J. Immunol. 194, 5465–5471 (2015)
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008)
Leemans, J. C. et al. Depletion of alveolar macrophages exerts protective effects in pulmonary tuberculosis in mice. J. Immunol. 166, 4604–4611 (2001)
Jia, W. & He, Y.-W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J. Immunol. 186, 5313–5322 (2011)
Malhotra, R., Warne, J. P., Salas, E., Xu, A. W. & Debnath, J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11, 145–154 (2015)
Komatsu, M. et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 (2007)
Lee, E.-J. & Tournier, C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7, 689–695 (2011)
McAlpine, F., Williamson, L. E., Tooze, S. A. & Chan, E. Y. W. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy 9, 361–373 (2013)
Fleming, T. J., Fleming, M. L. & Malek, T. R. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J. Immunol. 151, 2399–2408 (1993)
Acknowledgements
C.L.S. is supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation. J.M.K. is supported by a National Science Foundation Graduate Research Fellowship DGE-1143954 and the NIGMS Cell and Molecular Biology Training Grant GM007067. J.P.H. is supported by a National Science Foundation Graduate Research Fellowship DGE-1143954. H.W.V., S.P. and A.K. are supported by U19 AI109725. We would like to acknowledge D. Kreamalmeyer for assistance with the mouse colonies and T. Malek and L. D. Sibley for providing the 1A8 hybridoma.
Author information
Authors and Affiliations
Contributions
J.M.K. designed and performed experiments, analysed data, and wrote the manuscript. J.P.H. and L.A.W. performed experiments. S.P., A.K. and J.D. generated mouse strains. H.W.V. provided all mouse strains, analysed data and wrote the manuscript. C.L.S. designed experiments, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
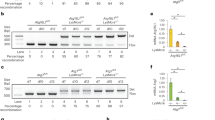
Extended Data Figure 1 Survival of mice with defects in autophagy genes other than Atg5.
Per cent survival of mice following infection with 100 colony-forming units (c.f.u.) of aerosolized M. tuberculosis. a, Survival of C57Bl/6 (open squares), Ulk1−/− (blue triangles), Ulk2−/− (inverted pink triangles), Atg4b−/− (red diamonds), and p62−/− (green circles) mice. b, Survival of Atg14lfl/fl-Lysm-cre (purple diamonds), Atg12fl/fl-Lysm-cre (red inverted triangles), Atg16l1fl/fl-Lysm-cre (green triangles), Atg7fl/fl-Lysm-cre (pink diamonds), Atg3fl/fl-Lysm-cre (brown circles) and corresponding floxed control mice. Floxed control mice are shown in open shapes, LysM–Cre-expressing mice are shown in closed shapes. c, Survival of C57Bl/6 (open squares), Atg16l1HM1 (open circles). Samples represent biological replicates. See Supplementary Fig. 6 for sample sizes.
Extended Data Figure 2 Analysis of autophagy in bronchoalveolar macrophages.
Western blot analysis of p62, LC3, and actin levels in ex vivo macrophages isolated from bronchoalveolar lavages of uninfected mice. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 Atg5fl/fl bone-marrow-derived macrophages are hypomorphic for ATG5.
Western blot analysis of ATG5 (ATG5–ATG12 conjugate, 56 kDa) and actin in uninfected-bone-marrow-derived macrophages. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4 Loss of Atg5 or Atg16l1 in LysM+ cells does not lead to increased c.f.u. at 2 w.p.i. log pulmonary c.f.u. at 2 w.p.i.
Samples represent biological replicates; error bars represent mean ± s.e.m. See Supplementary Fig. 7 for sample sizes and results from all statistical comparisons.
Extended Data Figure 5 Cytokine levels in uninfected lungs.
Concentration of cytokines in lungs (homogenized in 1 ml PBS plus 0.05% Tween 80) from uninfected mice. Levels of IFNγ, IL-6, MIP-1α, IL-17, and G-CSF were below the limit of detection. C57Bl/6 (grey solid bars), Atg5fl/fl (blue striped bars), Atg5fl/fl-Lysm-cre (blue solid bars), Atg16l1fl/fl (green striped bars), Atg16l1fl/fl-Lysm-cre (green solid bars). Statistical differences were determined by one-way ANOVA and Bonferonni’s multiple comparison test. n.s., not significant. Samples represent biological replicates; error bars represent mean ± s.e.m. See Supplementary Fig. 8 for sample sizes and results from all statistical comparisons.
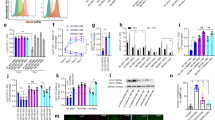
Extended Data Figure 6 Number of inflammatory cells in lungs of mice at 2 and 3 w.p.i. (related to Fig. 2).
a, Gating strategy for analysis of inflammatory cells in lungs at 2 and 3 w.p.i. Single lung cells were gated based on CD11b, CD11c, Ly6G, Ly6C and autofluorescence (auto). The parental gate is shown above each contour plot. Representative data are shown from an Atg5fl/fl mouse at 2 w.p.i. b, c, C57Bl/6 (grey solid bars), Atg5fl/fl (blue striped bars), Atg5fl/fl-Lysm-cre (blue solid bars), Atg16l1fl/fl (green striped bars), Atg16l1fl/fl-Lysm-cre (green solid bars). Mean number of alveolar macrophages, PMNs, recruited macrophages, and inflammatory monocytes in lungs at 2 w.p.i. (b) and 3 w.p.i. (c). d, e, Flow cytometry data presented in b and c and in Fig. 2 are the compilation of results from five experiments. In some experiments, different amounts of lung were collected for analysis, making it difficult to compare the average number of each cell type between strains, unless the data are normalized (as done in Fig. 2c, d—percentage of total cells). Therefore, to compare the raw number of cells detected in each cell population, each mouse analysed at 2 w.p.i. (d) and 3 w.p.i. (e) has been graphed individually. Each line represents a different mouse, with dots indicating the number of total cells, alveolar macrophages, PMNs, recruited macrophages and inflammatory monocytes. Statistical differences were determined by one-way ANOVA and Bonferonni’s multiple comparison test (b, c); *P < 0.05; n.s., not significant. Samples represent biological replicates; error bars represent mean ± s.e.m. See Supplementary Fig. 9 for sample sizes and results from all statistical comparisons.
Extended Data Figure 7 Number of inflammatory cells in lungs of mice at 3 w.p.i. (related to Fig. 4).
Number of alveolar macrophages, PMNs, recruited macrophages, and inflammatory monocytes in lungs at 3 w.p.i. C57Bl/6 (grey solid bars), Atg5fl/fl (blue striped bars), Atg5fl/fl-Lysm-cre (blue solid bars), ‘healthy’ Atg5fl/fl-MRP8-cre (purple striped bars), and ‘susceptible’ Atg5fl/fl-MRP8-cre (purple solid bars). Statistical differences were determined by one-way ANOVA and Bonferonni’s multiple comparison test; *P < 0.05; n.s., not significant. Samples represent biological replicates; error bars represent mean ± s.e.m. See Supplementary Fig. 10 for sample sizes and results from all statistical comparisons.
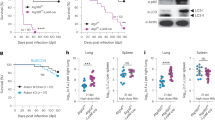
Extended Data Figure 8 Analysis of autophagy in bone marrow PMNs.
Western blot analysis of p62, LC3, and actin in bone marrow PMNs from uninfected mice. Each lane represents an individual mouse. Two replicates of the Atg5fl/fl-Lysm-cre and Atg16l1fl/fl-Lysm-cre mice are shown. For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Figure 1
This file contains source gel data showing uncropped western blot images and ladder markers. Boxes indicate how each image was cropped for final figures. (PDF 651 kb)
Rights and permissions
About this article
Cite this article
Kimmey, J., Huynh, J., Weiss, L. et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569 (2015). https://doi.org/10.1038/nature16451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16451
This article is cited by
-
Dimethyl itaconate is effective in host-directed antimicrobial responses against mycobacterial infections through multifaceted innate immune pathways
Cell & Bioscience (2023)
-
ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages
Nature Microbiology (2023)
-
Autophagy restricts Mycobacterium tuberculosis during acute infection in mice
Nature Microbiology (2023)
-
Autophagy is part of the answer to tuberculosis
Nature Microbiology (2023)
-
Immune evasion and provocation by Mycobacterium tuberculosis
Nature Reviews Microbiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.