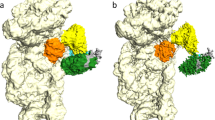
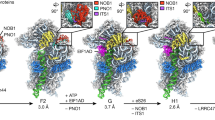
Abstract
During eukaryotic translation initiation, 43S complexes, comprising a 40S ribosomal subunit, initiator transfer RNA and initiation factors (eIF) 2, 3, 1 and 1A, attach to the 5′-terminal region of messenger RNA and scan along it to the initiation codon. Scanning on structured mRNAs also requires the DExH-box protein DHX29. Mammalian eIF3 contains 13 subunits and participates in nearly all steps of translation initiation. Eight subunits having PCI (proteasome, COP9 signalosome, eIF3) or MPN (Mpr1, Pad1, amino-terminal) domains constitute the structural core of eIF3, to which five peripheral subunits are flexibly linked. Here we present a cryo-electron microscopy structure of eIF3 in the context of the DHX29-bound 43S complex, showing the PCI/MPN core at ∼6 Å resolution. It reveals the organization of the individual subunits and their interactions with components of the 43S complex. We were able to build near-complete polyalanine-level models of the eIF3 PCI/MPN core and of two peripheral subunits. The implications for understanding mRNA ribosomal attachment and scanning are discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Cryo-EM maps and atomic coordinates for the reported structures have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-3056, EMD-3057 and EMD-3058, and in the Protein Data Bank under the accession numbers 5A5T and 5A5U.
References
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Rev. Mol. Cell Biol. 11, 113–127 (2010)
Hinnebusch, A. G. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562 (2006)
Valásek, L. S. ‘Ribozoomin’–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 13, 305–330 (2012)
Ellisdon, A. M. & Stewart, M. Structural biology of the PCI-protein fold. BioArchitecture 2, 118–123 (2012)
Pena, V., Liu, S., Bujnicki, J. M., Lührmann, R. & Wahl, M. C. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell 25, 615–624 (2007)
Enchev, R. I., Schreiber, A., Beuron, F. & Morris, E. P. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure 18, 518–527 (2010)
Hashem, Y. et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153, 1108–1119 (2013)
Hashem, Y. et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539–543 (2013)
Querol-Audi, J. et al. Architecture of human translation initiation factor 3. Structure 21, 920–928 (2013)
Sun, C. et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl Acad. Sci. USA 108, 20473–20478 (2011)
Beck, F. et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl Acad. Sci. USA 109, 14870–14875 (2012)
Lander, G. C. et al. Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 (2012)
Lingaraju, G. M. et al. Crystal structure of the human COP9 signalosome. Nature 512, 161–165 (2014)
ElAntak, L., Tzakos, A. G., Locker, N. & Lukavsky, P. J. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J. Biol. Chem. 282, 8165–8174 (2007)
Herrmannová, A. et al. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res. 40, 2294–2311 (2012)
Liu, Y. et al. Translation initiation factor eIF3b contains a nine-bladed β-propeller and interacts with the 40S ribosomal subunit. Structure 22, 923–930 (2014)
Erzberger, J. P. et al. Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex. Cell 158, 1123–1135 (2014)
Zhou, M. et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl Acad. Sci. USA 105, 18139–18144 (2008)
Dong, Z., Qi, J., Peng, H., Liu, J. & Zhang, J. T. Spectrin domain of eukaryotic initiation factor 3a is the docking site for formation of the a:b:i:g subcomplex. J. Biol. Chem. 288, 27951–27959 (2013)
Wagner, S., Herrmannová, A., Malík, R., Peclinovská, L. & Valášek, L. S. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol. Cell. Biol. 34, 3041–3052 (2014)
Masutani, M., Sonenberg, N., Yokoyama, S. & Imataka, H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 26, 3373–3383 (2007)
Fraser, C. S. et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J. Biol. Chem. 279, 8946–8956 (2004)
Valásek, L., Hasek, J., Trachsel, H., Imre, E. M. & Ruis, H. The Saccharomyces cerevisiae HCR1 gene encoding a homologue of the p35 subunit of human translation initiation factor 3 (eIF3) is a high copy suppressor of a temperature-sensitive mutation in the Rpg1p subunit of yeast eIF3. J. Biol. Chem. 274, 27567–27572 (1999)
Estrin, E., Lopez-Blanco, J. R., Chacon, P. & Martin, A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 21, 1624–1635 (2013)
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013)
Schmitt, E. et al. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nature Struct. Mol. Biol. 19, 450–454 (2012)
Dhote, V., Sweeney, T. R., Kim, N., Hellen, C. U. & Pestova, T. V. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl Acad. Sci. USA 109, E3150–E3159 (2012)
Aylett, C. H., Boehringer, D., Erzberger, J. P., Schaefer, T. & Ban, N. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nature Struct. Mol. Biol. 22, 269–271 (2015)
Kouba, T. et al. Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 subunit. PLoS ONE 7, e40464 (2012)
Valásek, L. et al. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 17, 786–799 (2003)
Pisarev, A. V., Kolupaeva, V. G., Yusupov, M. M., Hellen, C. U. & Pestova, T. V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 27, 1609–1621 (2008)
Zhou, C. et al. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 3, 14 (2005)
Rezende, A. M. et al. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates-identification of conserved and divergent features based on orthologue analysis. BMC Genomics 15, 1175 (2014)
LeFebvre, A. K. et al. Translation initiation factor eIF4G–1 binds to eIF3 through the eIF3E subunit. J. Biol. Chem. 281, 22917–22932 (2006)
Villa, N., Do, A., Hershey, J. W. B. & Fraser, C. S. Human eukaryotic initiation factor 4G (eIF4G) binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 288, 32932–32940 (2013)
Yu, Y., Abaeva, I. S., Marintchev, A., Pestova, T. V. & Hellen, C. U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 39, 4851–4865 (2011)
Chiu, W. L. et al. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 30, 4415–4434 (2010)
Elantak, L. et al. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol. 396, 1097–1116 (2010)
Cuchalová, L. et al. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 30, 4671–4686 (2010)
Pisareva, V. P., Pisarev, A. V., Komar, A. A., Hellen, C. U. & Pestova, T. V. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135, 1237–1250 (2008)
Grassucci, R. A., Taylor, D. J. & Frank, J. Preparation of macromolecular complexes for cryo-electron microscopy. Nature Protocols 2, 3239–3246 (2007)
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988)
Wagenknecht, T., Frank, J., Boublik, M., Nurse, K. & Ofengand, J. Direct localization of the tRNA–anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 203, 753–760 (1988)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Langlois, R. et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J. Struct. Biol. 186, 1–7 (2014)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Kucukelbir, A., Sigworth, S. J. & Tagare, D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010)
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226 (2015)
Mitchell, A. et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 (2015)
Marintchev, A. & Wagner, G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 37, 197–284 (2004)
Lange, H. et al. The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 10, e1004564 (2014)
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999)
Thiele, H., Glandorf, J. & Hufnagel, P. Bioinformatics strategies in life sciences: from data processing and data warehousing to biological knowledge extraction. J. Integr. Bioinform. 7, 141 (2010)
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 (2010)
Khoshnevis, S. et al. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res. 42, 4123–4139 (2014)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Wei, Z. et al. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J. Biol. Chem. 279, 34983–34990 (2004)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Hashem, Y. et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 494, 385–389 (2013)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 1, 33–8. 27–8 (1996)
Brooks, B. R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983)
MacKerell, A. D. Jr. et al. in The Encyclopedia of Computational Chemistry (eds von Rague Schleyer, P. et al.) 271–277 (John Wiley & Sons, 1998)
Acknowledgements
We thank R. A. Grassucci for helping collect the electron microscopy images, H. Kao for help with computations, and the proteomic platform Strasbourg-Esplanade for conducting the nano-LS–MS/MS analysis. This work was supported by both HHMI and NIH R01 GM29169 (to J.F.) and NIH R01 GM59660 (to T.V.P.). This work has been published under the framework of the LABEX: ANR-10-LABX-0036_NETRNA and benefits from funding from the state managed by the French National Research Agency as part of the Investments for the future program (to Y.H.).
Author information
Authors and Affiliations
Contributions
A.d.G., V.D., L.K., C.U.T.H., T.V.P., J.F. and Y.H. interpreted the data and wrote the manuscript. V.D. and T.V.P. prepared the samples. Y.H. and A.d.G. performed the cryo-electron microscopy experiments, data processing and the atomic modelling. L.K. performed the mass-spectrometry experiment. T.V.P., J.F. and Y.H. directed research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Domain organization of the eIF3 subunits and resolution of eIF3 core.
a, Schematic representation of the domain organization of the rabbit eIF3 subunits (see Methods). Domain boundaries are indicated, and based where possible on our polyalanine-level model of eIF3. HD, helical domain; Z, zinc-recognition motif. Dashed line in eIF3l subunit diagram indicates that the helical domain might extend further in the N terminus, but it was not possible to be conclusive based on our density map. b, Fourier shell correlation (FSC) of the different 43S complex reconstructions used during analysis. The resolution estimation followed the ‘gold standard’ protocol ensuring independence of the half-set reconstructions. x axis, resolution in Å; y axis, FSC. Green line denotes 43S complexes including particles that present the eIF3 core subunits; blue line denotes 43S complexes including all particles presenting DHX29 and the eIF3 peripheral subunits b, g and i; and red line denotes 43S complexes including all particles presenting the density attributed to eIF3d. For each reconstruction, a dashed line marks the resolution at which the FSC reaches the value of 0.143. c–f, Qualitative comparison of eIF3 core resolution in the present and in previous structures. c, d, eIF3 core cryo-EM structure from our previous study at 11.6 Å (ref. 7). e, f, eIF3 core cryo-EM structure from the present study at 6 Å after focused refinement. The eIF3 core is labelled according to the anthropomorphic nomenclature. g, Unassigned density region of the eIF3 core cryo-EM structure, coloured in red, seen from three different orientations. The green surface represents most of the core region that was modelled.
Extended Data Figure 2 Sorting of particle images and focused refinement of the five-lobed core of eIF3.
a, Composition of eIF3 purified from RRL for assembly of 43S complexes, resolved by SDS–PAGE and analysed by nano-LC–MS/MS to characterize truncation of eIF3a and eIF3c due to endoproteolytic cleavage. The intensity of labels in this panel reflects the intensity of bands corresponding to the truncated forms of eIF3a and eIF3c. The sequence of the N-terminal region of rabbit eIF3b has not been conclusively established (see Extended Data Fig. 9c) and numbering therefore refers to human eIF3b. b, Overview of the process of sorting particle images. The population of each class is indicated by the number of particles and the percentage of the total number of particles at the beginning of each of the two classification rounds. The DHX29-bound 43S complex was processed from a total of ∼650,000 particle images, which were first sorted into ten different classes. Class 2 (∼125,000 particles) was sorted into ten subclasses, which are displayed in four different orientations, showing the intersubunit face, front, solvent side and bottom, respectively. Classes 2–7 and 9 from the second classification round were pooled and refined yielding a reconstruction presenting a variable resolution ranging from 4.5 to 15 Å (bottom right). d, f, Cryo-EM reconstruction of the 43S complex, coloured according to the local resolution as measured using ResMap (see Methods). The red and black boxes correspond to close-up views of the eIF3 core viewed from the intersubunit face (d) and the solvent face (f) of the 40S subunit. c, e, CryoEM reconstruction of the eIF3 core after focused refinement, coloured according to the local resolution as measured using ResMap. Maps in c–f are filtered to 6 Å.
Extended Data Figure 3 Comparison of the structure of the mammalian eIF3 core with the structures of the COP9 signalosome and 26S proteasome lid molecules.
a–c, Close-up views of the helical bundles of eIF3, COP9 and 26S lid molecules. d–i, All three molecules shown in two different orientations. Different constitutive subunits are labelled and coloured variably. Homologous subunits among all three molecules are shaded using the same colour in all panels. j, k, Additional quaternary interactions between eIF3a and eIF3c, and between eIF3c and eIF3e, which occur neither in COP9 nor in 26S lid molecules. l, Consequence of these additional quaternary interactions on the structure of eIF3 subunits a, c and e, schematized as rectangles. Black arrows around axes describe the rotation of the helical domains of subunits a, c and e relatively to their respective PCI domains, owing to the existence of these additional quaternary interactions, compared to COP9 and 26S lid molecules.
Extended Data Figure 4 Conservation of quaternary interactions between eIF3a and eIF3c subunits, and between eIF3c and eIF3e subunits.
a, Ribbon representation of eIF3a and eIF3c subunits. b–d, Close-up views of contact regions between eIF3a and eIF3c. e, Ribbon representation of eIF3 c and e subunits. f, g, Close-up views of contact regions between subunits c and e of eIF3. Red spheres represent residues at the interfaces that are conserved in eIF3 from six representative eukaryotic organisms; H. sapiens, C. elegans, A. thaliana, D. melanogaster and X. tropicalis, which are very different multicellular eukaryotic organisms, and N. crassa, a unicellular organism. These organisms all have a full complement of 13 eIF3 subunits. Orange spheres represent residues at the interfaces that are conserved only in the five multicellular eukaryotic organisms. The remaining residues that are suggested by the model and the density map to be involved in quaternary interactions are represented as ribbons in salmon colour. Many of these other residues are conserved in three or four of the compared organisms, but almost all of them have conserved chemical properties.
Extended Data Figure 5 Conservation of quaternary interactions between eIF3 subunits a, m, f and h.
a, In centre, eIF3 a, m, f and h subunits. b–f, Close-up views of contact regions between subunits a, m, f and h of eIF3. Red spheres represent residues at the interfaces that are conserved in six representative eukaryotic organisms; H. sapiens, C. elegans, A. thaliana, D. melanogaster, X. tropicalis and the unicellular N. crassa. Orange spheres represent residues at the interfaces that are conserved only in the five multicellular eukaryotic organisms. The remaining residues that are suggested by the model and the density map to be involved in quaternary interactions are represented as ribbons in salmon colour. Many of these other residues are conserved in three or four of the compared organisms, but almost all of them have conserved chemical properties.
Extended Data Figure 6 Conservation of quaternary interactions between eIF3 subunits e, k and l, and between eIF3 subunits in the region of the helical bundle.
a, Ribbon representation of eIF3 e, k and l subunits. b–f, Close-up views of contact regions between subunits e, k and l of eIF3. g, Ribbon representation of eIF3 octamer core. h, Close-up views of contact regions between subunits a, c, e, m, f, h, k and l of eIF3 in the helical bundle region, seen from the direction of the axis of the latter. i, Region displayed in b rotated by 90°. Red spheres represent residues at the interfaces that are conserved in six representative eukaryotic organisms; H. sapiens, C. elegans, A. thaliana, D. melanogaster, X. tropicalis and the unicellular N. crassa. Orange spheres represent residues at the interfaces that are conserved only in the five multicellular eukaryotic organisms. The remaining residues that are suggested by the model and the density map to be involved in quaternary interactions are represented as ribbons in salmon colour. Many of these other residues are conserved in three or four of the compared organisms, but almost all of them have conserved chemical properties. j, Same view as in i displaying all the hydrophobic residues of the helical bundle region in silver ribbons. The abundance of hydrophobic residues in the helical bundle at the interfaces of different helices suggests the stabilization of the bundle though hydrophobic interactions, hence the low identity conservation of residues at the interfaces.
Extended Data Figure 7 Comparison of the mammalian eIF3 core model built from the 6 Å cryo-EM reconstruction with a model based on low-resolution cryo-EM studies.
In the centre, our polyalanine-level model of the mammalian eIF3 octamer core (represented in red ribbons) fitted on the atomic model proposed previously18 (represented in dark grey ribbons), shown in two different orientations. The surrounding panels are close-up views of different constitutive subunits, highlighting notable structural differences between the previously proposed model18 and the model proposed in this study. Gold- and cyan-dashed ovals highlight the misassignment of several helices of the helical bundle belonging to the C termini of subunits h and f, respectively, in the previously proposed model. Red-, green-dashed circles and black arrowhead highlight the absence in the previously proposed model of important structural features involved in quaternary interactions. In each panel, the remaining subunits of the eIF3 core octamer are faded out as transparent ribbons.
Extended Data Figure 8 Sorting of particle images and focused classification of eIF3 peripheral subunits near the mRNA channel entrance and exit.
a, Overview of the process of sorting particle images. The population of each class is indicated by the number of particles and the percentage of the total number of particles at the beginning of each of the classification rounds. After the first round of classification, class 2 stands out as the class displaying eIF3. Focused classification of eIF3 peripheral subunits, near the mRNA channel entrance, was performed by applying a smooth-edge mask corresponding to the shape of the concerned subunits of eIF3 and to a region of the 43S complex encompassing DHX29 and h16 of the 40S subunit. The mask is displayed as pink mesh. The resulting classes from the focused sorting of class 2 of the first classification round are displayed in two different orientations, front and solvent side, respectively. Class 6 from the second classification round presents the most solid and complete density of the peripheral subunits of eIF3 at this region of the complex, and it was therefore refined yielding a reconstruction presenting an average resolution of 7.1 Å. Cryo-EM reconstruction of eIF3b and eIF3i along with DHX29, coloured by local resolution. b, After the first round of classification, on class 2, focused classification of an eIF3 peripheral subunit, identified as eIF3d, near the mRNA channel exit behind ribosomal protein RACK1, was performed by applying a smooth-edge mask corresponding to the shape of the concerned subunit of eIF3 and to a region of the head of the 40S subunit encompassing RACK1. The mask is displayed as pink mesh. The resulting classes from the focused sorting of class 2 of the first classification round are displayed in two different orientations, intersubunit face and top, respectively. Class 2 from the second round of classification presents the most solid and complete density of the peripheral subunit of eIF3 at this region of the complex, but due to some apparent heterogeneity in eIF3d, a third classification round was required, yielding four classes displaying a solid eIF3d subunit in slightly different conformations (other classes obtained in this third round of classification were completely empty and therefore not shown). The major class (39% of the particles in this round) yielded a reconstruction presenting an average resolution of 7.7 Å. Cryo-EM reconstruction of eIF3d, coloured by local resolution.
Extended Data Figure 9 Shape and ribosomal binding site of eIF3d, and eIF3b sequence.
a, Segmented cryo-EM reconstruction of the peripheral eIF3d subunit localized on the head of the 40S subunit, behind ribosomal protein RACK1, displayed at a high density threshold to show its most solid features, in four different orientations. b, eIF3d subunit in the context of the 43S preinitiation complex, seen from the back, showing the ribosomal proteins that contact it directly. This figure is complementary to Fig. 4c as it displays the same complex in a different orientation. This panel shows contacts between eIF3d subunit and ribosomal proteins eS28, uS7 and uS9. Contacts with RACK1 cannot be seen from this orientation. c, eIF3b sequence. The amino acid sequences of human eIF3b (GenBank NP_003742.2) and rabbit eIF3b (UniProt G1SZ03_RABIT) aligned using T_COFFEE (http://www.ebi.ac.uk/Tools/msa/tcoffee/) and annotated to show identity with tryptic and chymotryptic peptides derived from purified rabbit eIF3 and identified by nano-LC–MS/MS analysis. The complete sequence of rabbit eIF3b has not been determined, but clearly extends beyond the N terminus of G1SZ03_RABIT, and we therefore used the numbering of residues in human eIF3b when referring in the text to elements of rabbit eIF3b.
Supplementary information
Supplementary Information
This file contains Supplementary Text. (PDF 101 kb)
Rights and permissions
About this article
Cite this article
des Georges, A., Dhote, V., Kuhn, L. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525, 491–495 (2015). https://doi.org/10.1038/nature14891
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14891
This article is cited by
-
The molecular basis of translation initiation and its regulation in eukaryotes
Nature Reviews Molecular Cell Biology (2024)
-
The deubiquitinase EIF3H promotes hepatocellular carcinoma progression by stabilizing OGT and inhibiting ferroptosis
Cell Communication and Signaling (2023)
-
Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3–eIF3h interaction
Molecular Cancer (2022)
-
Dynamic eIF3a O-GlcNAcylation controls translation reinitiation during nutrient stress
Nature Chemical Biology (2022)
-
METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis
Nature Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.