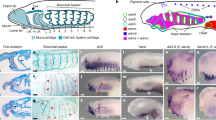
Abstract
The sudden appearance of the neural crest and neurogenic placodes in early branching vertebrates has puzzled biologists for over a century1. These embryonic tissues contribute to the development of the cranium and associated sensory organs, which were crucial for the evolution of the vertebrate “new head”2,3. A previous study suggests that rudimentary neural crest cells existed in ancestral chordates4. However, the evolutionary origins of neurogenic placodes have remained obscure owing to a paucity of embryonic data from tunicates, the closest living relatives to those early vertebrates5. Here we show that the tunicate Ciona intestinalis exhibits a proto-placodal ectoderm (PPE) that requires inhibition of bone morphogenetic protein (BMP) and expresses the key regulatory determinant Six1/2 and its co-factor Eya, a developmental process conserved across vertebrates. The Ciona PPE is shown to produce ciliated neurons that express genes for gonadotropin-releasing hormone (GnRH), a G-protein-coupled receptor for relaxin-3 (RXFP3) and a functional cyclic nucleotide-gated channel (CNGA), which suggests dual chemosensory and neurosecretory activities. These observations provide evidence that Ciona has a neurogenic proto-placode, which forms neurons that appear to be related to those derived from the olfactory placode and hypothalamic neurons of vertebrates. We discuss the possibility that the PPE-derived GnRH neurons of Ciona resemble an ancestral cell type, a progenitor to the complex neuronal circuit that integrates sensory information and neuroendocrine functions in vertebrates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gaskell, W. H. The Origin of Vertebrates (Longmans, Green & Co., 1908)
Steventon, B., Mayor, R. & Streit, A. Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol. 389, 28–38 (2014)
Gans, C. & Northcutt, R. G. Neural crest and the origin of vertebrates: a new head. Science 220, 268–273 (1983)
Abitua, P. B., Wagner, E., Navarrete, I. A. & Levine, M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492, 104–107 (2012)
Delsuc, F., Brinkmann, H., Chourrout, D. & Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006)
Shimeld, S. M. & Holland, P. W. Vertebrate innovations. Proc. Natl Acad. Sci. USA 97, 4449–4452 (2000)
Patthey, C., Schlosser, G. & Shimeld, S. M. The evolutionary history of vertebrate cranial placodes – I: cell type evolution. Dev. Biol. 389, 82–97 (2014)
Mazet, F. et al. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 282, 494–508 (2005)
Christiaen, L., Bourrat, F. & Joly, J. A modular cis-regulatory system controls isoform-specific pitx expression in ascidian stomodæum. Dev. Biol. 277, 557–566 (2005)
Sutton, S. W. et al. Distribution of G-protein-coupled receptor (GPCR) 135 binding sites and receptor mRNA in the rat brain suggests a role for relaxin-3 in neuroendocrine and sensory processing. Neuroendocrinology 80, 298–307 (2004)
Schwanzel-Fukuda, M. & Pfaff, D. W. Origin of luteinizing hormone-releasing hormone neurons. Nature 338, 161–164 (1989)
Wray, S., Grant, P. & Gainer, H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl Acad. Sci. USA 86, 8132–8136 (1989)
Kwon, H.-J., Bhat, N., Sweet, E. M., Cornell, R. A. & Riley, B. B. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 6, e1001133 (2010)
Ruf, R. G. et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1–SIX1-DNA complexes. Proc. Natl Acad. Sci. USA 101, 8090–8095 (2004)
Reichert, S., Randall, R. A. & Hill, C. S. A BMP regulatory network controls ectodermal cell fate decisions at the neural plate border. Development 140, 4435–4444 (2013)
Wagner, E. & Levine, M. FGF signaling establishes the anterior border of the Ciona neural tube. Development 139, 2351–2359 (2012)
Ahrens, K. & Schlosser, G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis . Dev. Biol. 288, 40–59 (2005)
Wray, S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J. Neuroendocrinol. 22, 743–753 (2010)
Kusakabe, T. G. et al. A conserved non-reproductive GnRH system in chordates. PLoS ONE 7, e41955 (2012)
McGowan, B. M. et al. Relaxin-3 stimulates the hypothalamic-pituitary-gonadal axis. Am. J. Physiol. Endocrinol. Metab. 295, E278–E286 (2008)
Terakado, K. Induction of gamete release by gonadotropin-releasing hormone in a protochordate, Ciona intestinalis . Gen. Comp. Endocrinol. 124, 277–284 (2001)
Konno, A. et al. Distribution and structural diversity of cilia in tadpole larvae of the ascidian Ciona intestinalis . Dev. Biol. 337, 42–62 (2010)
Reese, T. S. Olfactory cilia in the frog. J. Cell Biol. 25, 209–230 (1965)
Kaupp, U. B. Olfactory signalling in vertebrates and insects: differences and commonalities. Nature Rev. Neurosci. 11, 188–200 (2010)
Constantin, S. & Wray, S. Gonadotropin-releasing hormone-1 neuronal activity is independent of cyclic nucleotide-gated channels. Endocrinology 149, 279–290 (2008)
El-Majdoubi, M. & Weiner, R. I. Localization of olfactory cyclic nucleotide-gated channels in rat gonadotropin-releasing hormone neurons. Endocrinology 143, 2441–2444 (2002)
Boehm, U., Zou, Z. & Buck, L. B. Feedback loops link odor and pheromone signaling with reproduction. Cell 123, 683–695 (2005)
Yoon, H., Enquist, L. W. & Dulac, C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123, 669–682 (2005)
Gorbman, A. Olfactory origins and evolution of the brain-pituitary endocrine system: facts and speculation. Gen. Comp. Endocrinol. 97, 171–178 (1995)
Arendt, D. The evolution of cell types in animals: emerging principles from molecular studies. Nature Rev. Genet. 9, 868–882 (2008)
Christiaen, L., Stolfi, A. & Levine, M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 340, 179–187 (2010)
Rehm, E. J., Hannibal, R. L., Chaw, R. C., Vargas-Vila, M. A. & Patel, N. H. Fixation and dissection of Parhyale hawaiensis embryos. Cold Spring Harb Protoc. 2009, pdb.prot5127 (2009)
Hudson, C. & Yasuo, H. Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development 132, 1199–1210 (2005)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013)
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014)
Kamesh, N., Aradhyam, G. K. & Manoj, N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis . BMC Evol. Biol. 8, 129 (2008)
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007)
Winchell, C. J. & Jacobs, D. K. Expression of the Lhx genes apterous and lim1 in an errant polychaete: implications for bilaterian appendage evolution, neural development, and muscle diversification. EvoDevo 4, 4 (2013)
Isberg, V. et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 42, D422–D425 (2014)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009)
Dhallan, R. S., Yau, K. W., Schrader, K. A. & Reed, R. R. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347, 184–187 (1990)
Hotta, K. et al. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 236, 1790–1805 (2007)
Acknowledgements
We thank Y. Miyamoto and M. Kotera for technical assistance and A. Stolfi for cloning Chordin>GFP. This work was supported by a grant from the National Institutes of Health (NS076542) and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (25650118, 25290067) and from the Japan Space Forum (h160179). Portions of this study were facilitated by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology in Japan. The work of C.H. in the laboratory of H. Yasuo was funded by the Agence Nationale de la Recherche (ANR-09-BLAN-0013-01). P.B.A. and A.N.K. were supported by predoctoral fellowships from the National Science Foundation and California Institute for Regenerative Medicine, respectively.
Author information
Authors and Affiliations
Contributions
P.B.A. designed and performed most experiments in consultation with M.L. T.B.G. performed the Six1/2 and Eya in situ hybridizations. A.N.K. performed the larval colorimetric in situ hybridizations. C.J.W. performed the phylogenetic analysis. C.H. performed the BMP2/4 and Chordin in situ hybridizations. T.G.K. identified and cloned Ciona CNGs and GnRHs and analysed their expression. T.G.K., M.N., K.K. and M.T. performed functional analysis of CNGs.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Lineage information for Six1/2 expression in Ciona intestinalis from the gastrula to initial tailbud stage.
a, Schematic of the anterior neural plate border at the mid-gastrula stage, including cell lineage nomenclature. Dmrt is initially activated in six blastomeres of 64-cell embryos (a7.9, a7.10 and a7.13). This lineage produces only the anterior neural plate (and the adjacent anterior neural plate border), which forms the PPE. The anterior-most ZicL + cells of row IV (yellow) mark the boundary of the neural plate, which gives rise to the anterior sensory vesicle in tailbud embryos. The dotted line indicates the oral opening. b, Schematic of the anterior neural plate border during the gastrula–neurula transition, indicating the lineage-specific expression of Six1/2 (magenta). Six1/2 is initially expressed in eight cells comprising the posterior cells (row V) and the posterior lateral cells (row VI). c, Dorsal view of an initial tailbud embryo co-electroporated with Six1/2>mCherry, Dmrt>GFP and ZicL>CFP. At this stage, the cells initially expressing Six1/2 have divided once, giving rise to 16 cells in total. Brackets indicate the derivatives of the annotated lineages shown in b. Anterior is to the left.
Extended Data Figure 2 Six1/2 + cell morphogenesis in Ciona intestinalis from the initial tailbud stage to late tailbud I.
a–c, Dorsal views of tailbud embryos electroporated with Six1/2>mCherry (magenta) and counterstained with phalloidin (blue). a, At the initial tailbud stage I (according to Hotta et al.44), the Six1/2 + cells are arranged in a U shape, anterior to the neural plate. a′–c′, Underlined cell lineages are derived from the left side of the embryo. a′, Schematic indicates that the U shape is composed of 16 cells in total at the 11th generation (that is, a11.154, a11.138, etc.). There are no further divisions of these cells until after the late tailbud I stage. The green cells indicate a11.205, which are fated to become PPE-derived GnRH neurons. b, At the initial tailbud stage II, the lateral edges of the Six1/2 + cells begin to intercalate towards the midline. b′, The schematic shows a dotted circle where the future opening of the oral siphon forms. c, At the late tailbud I stage, the Six1/2 + cells have completed intercalation. The bright phalloidin signal in the centre of the pattern marks the apically localized actin of cells constricted towards the oral opening. The arrowheads indicate a PPE-derived cell fated to become a GnRH neuron. At this stage, the Six1/2 + cells are positioned on top of the anterior sensory vesicle over the ocellus. c′, The schematic shows a dotted circle where the oral opening forms. More anterior cells have undergone local cellular rearrangements. d–d″, Anterior lateral view of late tailbud I stage embryo electroporated with Six1/2>GFP and CNGA>mCherry. Asterisks indicate the ocellus. The arrowheads indicate a PPE-derived cell fated to become a GnRH neuron. d, Shows the Six1/2>GFP channel. d′, Shows the CNGA>mCherry channel (see Fig. 3c for larval expression). d″, Shows the merged image of Six1/2>GFP and CNGA>mCherry.
Extended Data Figure 3 Dorsal–ventral BMP patterning during PPE specification in Ciona intestinalis.
a, Schematic of the anterior neural plate border during the gastrula–neurula transition. Cell lineage nomenclature is used. Chordin and Six1/2 are co-expressed in the lateral posterior derivatives of rows V and VI (dark blue; also see Fig. 1b). b, Ventral view of mid-gastrula stage embryo hybridized with a BMP2/4 mRNA probe and merged with the nuclear counterstain 4′,6-diamidino-2-phenylindole (DAPI). c, Lateral view of an embryo during the gastrula–neurula transition hybridized with a BMP2/4 mRNA probe. Anterior is to the left. d, Dorsal view of an embryo during the gastrula–neurula transition hybridized with a Chordin mRNA probe and merged with a DAPI nuclear counterstain.
Extended Data Figure 4 Endogenous expression of newly described reporter genes in hatched Ciona larvae.
a–d, Bright field anterior lateral views. a, Animal hybridized with a GnRH2 mRNA probe. White arrowhead indicates the position of the oral opening. b, Animal hybridized with an RXFP3 mRNA probe. c, Animal hybridized with a SOG/Chemokine-like mRNA probe. The heavily stained tunic was manually removed to reveal the expression signal. d, Animal hybridized with a CNGA mRNA probe. Red arrows mark areas of comparable expression throughout the panels in presumed PPE-derived neurons. An adjacent signal in the ocellus-associated photoreceptors makes it difficult to discriminate expression in PPE-derived neurons in panel d.
Extended Data Figure 5 GnRH expression requires BMP attenuation.
a–d, Lateral view of a larva electroporated with GnRH>YFP and counterstained with phalloidin (violet). a, Larva co-electroporated with Dmrt>BMP2/4. Of the 200 larvae, 196 had no GnRH>YFP expression in aATENs and displayed mild to severe morphogenetic defects. Bracket shows mild anterior morphological defect. b, Larva co-electroporated with Dmrt>BMP5/7. Of the 200 larvae, 124 had GnRH>YFP expression in aATENs. c, Larva co-electroporated with Dmrt>BMPR1CA . Of the 200 larvae, 197 larvae had no GnRH>YFP expression in aATENs. d, Larva co-electroporated with Dmrt>TGF-βRCA . Of the 200 larvae, 130 had GnRH>YFP expression in aATENs and most displayed severe anterior neural tube defects. Arrowheads in b and d indicate the position of GnRH expression in aATENs. Anterior is to the left in all images.
Extended Data Figure 6 Initial phylogenetic analysis of two GPCRs expressed in the PPE-derived neurons of Ciona intestinalis.
This broad survey tree, constructed according to maximum likelihood, shows the approximate placement of the two GPCRs of interest (boxed in blue) within the rhodopsin-class G-protein-coupled receptors. The numbered Ciona sequences and their receptor-type identifications in parentheses are from Kamesh et al.37. All non-Ciona sequences were downloaded in bulk from http://pfam.xfam.org; they comprise the Pfam ‘seed’ alignment of seven-transmembrane receptors for the GPCRs. The rhodopsin subclass names are given to the right of each coloured group. The asterisk and double asterisk indicate the genes labelled RXFP3 and SOG/Chemokine-like, respectively, in Fig. 2e. We judge robust nodal support as bootstrap percentages >70 for minimum evolution (ME) and maximum likelihood (ML) and posterior probability percentages >95 for Bayesian inference (BI).
Extended Data Figure 7 Refined maximum likelihood phylogeny of two GPCRs expressed in the PPE-derived neurons of Ciona intestinalis.
Relative to the initial survey tree (see Extended Data Fig. 6), analysis of this focused set of ‘chemokine cluster’ sequences further clarifies the phylogenetic affinities of two Ciona GPCRs expressed in the aATEN–GnRH neurons. The latter sequences are highlighted with violet boxes; all other Ciona sequences are from Kamesh et al.37. The three nodal support values are (in order): ME bootstrap percentage, ML bootstrap percentage and BI posterior probability (only values >50 are shown). Branch lengths are proportional to molecular change (amino acid substitutions per site) between nodes; see scale bar for measurement.
Extended Data Figure 8 Elevation of intracellular Ca2+ concentration in cells expressing Ciona CNG channels in response to cyclic nucleotides.
a, Intracellular Ca2+ was visualized by Fura-2 ratiometric calcium imaging. Ca2+ influx into HEK 293 cells transfected with CNGA and CNGC was induced by 10 mM 8-Br-cGMP and 1 mM 8-Br-cAMP. In contrast, no Ca2+ influx was observed when HEK 293 cells transfected with CNGB were treated with either 10 mM 8-Br-cGMP or 1 mM 8-Br-cAMP. Coloured numbers in the ‘Before’ panels indicate cells that were subjected to a quantitative measurement of fluorescence. b, Dose-dependent response of HEK 293 cells transfected with CNGA to 8-Br-cAMP. Data were obtained from at least seven different cells in each of 16 different transfections. c, Dose-dependent response of HEK 293 cells transfected with CNGC to 8-Br-cAMP. Data were obtained from at least seven different cells in each of seven different transfections.
Supplementary information
Six1/2 expression in a neurula stage embryo
This video shows the neurulation of an embryo co-electroporated with Dmrt>H2B:YFP and Six1/2>mCherry from a dorsal view. The time-lapse covers a period of about 120 minutes, during which time Six1/2>mCherry becomes expressed in posterior row V cells. The video pauses to show the derivatives of rows III-IV (orange) and rows V-VI (white) at the 10th generation (i.e., a10.97, a10.98, etc.). Six1/2>mCherry is initially expressed in the derivatives of a10.77, a10.69, a10.101, and a10.103. This embryo was previously analyzed to show an extra cell division in the pigment cell lineage (Haupaix, N. et al. Ephrin-mediated restriction of ERK1/2 activity delimits the number of pigment cells in the Ciona CNS. Dev. Biol. 394, 170–180 (2014)). (MP4 16375 kb)
Co-expression of reporter genes in PPE-derived neurons
This video shows lateral views of the co-localization of SOG/Chemokine receptor-like>mCherry, RXFP3>mCherry, and CNGA>mCherry with GnRH>GFP in three larval stage embryos respectively. In each larva the mCherry reporter is shown first, as the plane of the Z-section pans through the left and right side of the animal. Then the GnRH>GFP signal is gradually faded in to show co-localization in the PPE-derived neurons. These neurons are located behind the oral opening (see Fig. 2c, 2d, and 3c) and are the only site of co-expression. (MP4 18735 kb)
Rights and permissions
About this article
Cite this article
Abitua, P., Gainous, T., Kaczmarczyk, A. et al. The pre-vertebrate origins of neurogenic placodes. Nature 524, 462–465 (2015). https://doi.org/10.1038/nature14657
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14657
This article is cited by
-
Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates
Nature Ecology & Evolution (2024)
-
Highly conserved and extremely evolvable: BMP signalling in secondary axis patterning of Cnidaria and Bilateria
Development Genes and Evolution (2024)
-
BMP signaling is required to form the anterior neural plate border in ascidian embryos
Development Genes and Evolution (2023)
-
Highly distinct genetic programs for peripheral nervous system formation in chordates
BMC Biology (2022)
-
Siphon-Specific Expression of an Actin Encoding Gene Is Regulated by Six1/2 in Ciona savignyi
Journal of Ocean University of China (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.