Abstract
Mitochondria have a major role in energy production via oxidative phosphorylation1, which is dependent on the expression of critical genes encoded by mitochondrial (mt)DNA. Mutations in mtDNA can cause fatal or severely debilitating disorders with limited treatment options2. Clinical manifestations vary based on mutation type and heteroplasmy (that is, the relative levels of mutant and wild-type mtDNA within each cell)3,4. Here we generated genetically corrected pluripotent stem cells (PSCs) from patients with mtDNA disease. Multiple induced pluripotent stem (iPS) cell lines were derived from patients with common heteroplasmic mutations including 3243A>G, causing mitochondrial encephalomyopathy and stroke-like episodes (MELAS)5, and 8993T>G and 13513G>A, implicated in Leigh syndrome. Isogenic MELAS and Leigh syndrome iPS cell lines were generated containing exclusively wild-type or mutant mtDNA through spontaneous segregation of heteroplasmic mtDNA in proliferating fibroblasts. Furthermore, somatic cell nuclear transfer (SCNT) enabled replacement of mutant mtDNA from homoplasmic 8993T>G fibroblasts to generate corrected Leigh-NT1 PSCs. Although Leigh-NT1 PSCs contained donor oocyte wild-type mtDNA (human haplotype D4a) that differed from Leigh syndrome patient haplotype (F1a) at a total of 47 nucleotide sites, Leigh-NT1 cells displayed transcriptomic profiles similar to those in embryo-derived PSCs carrying wild-type mtDNA, indicative of normal nuclear-to-mitochondrial interactions. Moreover, genetically rescued patient PSCs displayed normal metabolic function compared to impaired oxygen consumption and ATP production observed in mutant cells. We conclude that both reprogramming approaches offer complementary strategies for derivation of PSCs containing exclusively wild-type mtDNA, through spontaneous segregation of heteroplasmic mtDNA in individual iPS cell lines or mitochondrial replacement by SCNT in homoplasmic mtDNA-based disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
McBride, H. M., Neuspiel, M. & Wasiak, S. Mitochondria: more than just a powerhouse. Curr. Biol. 16, 551–560 (2006)
Wallace, D. C. A mitochondrial bioenergetic etiology of disease. J. Clin. Invest. 123, 1405–1412 (2013)
Taylor, R. W. & Turnbull, D. M. Mitochondrial DNA mutations in human disease. Nature Rev. Genet. 6, 389–402 (2005)
Grossman, L. I. & Shoubridge, E. A. Mitochondrial genetics and human disease. BioEssays 18, 983–991 (1996)
Goto, Y., Nonaka, I. & Horai, S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 (1990)
Tuppen, H. A., Blakely, E. L., Turnbull, D. M. & Taylor, R. W. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta 1797, 113–128 (2010)
Tachibana, M. et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature 493, 627–631 (2013)
Morten, K. J., Poulton, J. & Sykes, B. Multiple independent occurrence of the 3243 mutation in mitochondrial tRNAleuUUR in patients with the MELAS phenotype. Hum. Mol. Genet. 4, 1689–1691 (1995)
Chol, M. et al. The mitochondrial DNA G13513A MELAS mutation in the NADH dehydrogenase 5 gene is a frequent cause of Leigh-like syndrome with isolated complex I deficiency. J. Med. Genet. 40, 188–191 (2003)
Fujikura, J. et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia 55, 1689–1698 (2012)
Hamalainen, R. H. et al. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl Acad. Sci. USA 110, E3622–E3630 (2013)
Folmes, C. D. et al. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells 31, 1298–1308 (2013)
Picard, M. et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl Acad. Sci. USA 111, E4033–E4042 (2014)
Liu, G. H. et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491, 603–607 (2012)
Lian, X. et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nature Protocols 8, 162–175 (2013)
Tachibana, M. et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238 (2013)
Borchin, B., Chen, J. & Barberi, T. Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Rev. 1, 620–631 (2013)
Wallace, D. C., Brown, M. D. & Lott, M. T. Mitochondrial DNA variation in human evolution and disease. Gene 238, 211–230 (1999)
Brandon, M. C. et al. MITOMAP: a human mitochondrial genome database–2004 update. Nucleic Acids Res. 33, D611–D613 (2005)
Burgstaller, J. P. et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 7, 2031–2041 (2014)
Chinnery, P. F. et al. The challenges of mitochondrial replacement. PLoS Genet. 10, e1004315 (2014)
Folmes, C. D., Dzeja, P. P., Nelson, T. J. & Terzic, A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11, 596–606 (2012)
Henchcliffe, C. & Beal, M. F. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat. Clin. Pract. Neurol. 4, 600–609 (2008)
Boland, M. L., Chourasia, A. H. & Macleod, K. F. Mitochondrial dysfunction in cancer. Front. Oncol. 3, 292 (2013)
Mercer, T. R. et al. The human mitochondrial transcriptome. Cell 146, 645–658 (2011)
Ma, H. et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183 (2014)
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009)
Paull, D. et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 493, 632–637 (2013)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, (2014)
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnol. 28, 495–501 (2010)
Acknowledgements
The authors acknowledge the OHSU Embryonic Stem Cell Research Oversight Committee and the Institutional Review Board for providing oversight and guidance. We thank skin and oocyte donors and the Women's Health Research Unit staff at the Center for Women's Health, University Fertility Consultants and the Reproductive Endocrinology & Infertility Division in the Department of Obstetrics & Gynecology of Oregon Health & Science University for their support and procurement of gametes. We are grateful to M. Tachibana and A. Polat for help with derivation of PSCs and to M. Sparman for technical support. We are indebted to S. Gokhale for teratoma analysis and M. C. T. Penedo for microsatellite genotyping. We thank the staff at the Institute for Genomic Medicine Genomics Facility at UCSD for sequencing the RNA-seq libraries. Studies were supported by the Leducq Foundation, Mayo Clinic Center for Regenerative Medicine and OHSU and UCSD institutional funds. Work in the laboratory of J.C.I.B. was supported by the G. Harold and Leila Y. Mathers Charitable Foundation and the Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002).
Author information
Authors and Affiliations
Contributions
H.M. and S.M. conceived the study and designed the experiments. S.M., P.A., H.M., R.A., E.K., Y.L., N.M.G. and R.T.-H. derived and cultured PSCs. J.P. derived MELAS fibroblasts. H.M., T.H., Y.L., C.V.D., A.K. and N.M. performed the DNA/RNA extractions and mtDNA ARMs-qPCR analyses. C.D.L.F. and A.T. performed Seahorse studies on differentiated fibroblasts and data analysis. X.W. and T.H. performed MiSeq studies. R.M., S.M.-C. and L.C.L. performed RNA-seq and bioinformatic analysis of the data. J.W., A.O., L.M. and J.C.I.B. performed NPC, skeletal muscle cell, and cardiomyocyte differentiations, Seahorse studies and data analysis. H.M., C.D.L.F., J.W., R.M., D.P.W., L.C.L., A.T., J.C.I.B. and S.M. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
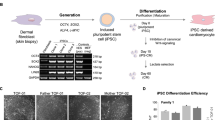
Extended Data Figure 1 mtDNA genotyping by Sanger sequencing.
a, Chromatographs showing mtDNA genotyping at 3243 position (arrow) in representative MELAS iPS cells. b, Chromatographs showing mtDNA genotyping at 8993 position (arrow). c, mtDNA at 13513 position (arrow) in representative iPS cells derived from Leigh syndrome patients. d, Chromatographs showing either wild-type A or mutant G allele at position 3243 in representative MELAS fibroblast clones. e, mtDNA genotyping demonstrated that all Leigh-iPS cell lines and Leigh-fib contain a G mutation allele at mtDNA position 8993. f, mtDNA genotyping demonstrated that Leigh-fib and Leigh-iPS1 cell lines contained a C mutant allele at position 4216 and a G mutant allele at position 8993, while Leigh-NT1 line carried oocyte mtDNA with a wild-type T allele at both positions.
Extended Data Figure 2 Cytogenetic, pluripotency and teratoma analyses.
a, MELAS-iPS1 and MELAS-iPS2 expressing NANOG detected by immunocytochemistry. Scale bars, 200 μm. b, Histological analyses of teratoma tumours produced after injections of MELAS-iPS1 and MELAS-iPS2 cells into SCID mice. Scale bars, 200 μm. c, Cytogenetic G-banding analysis confirmed that Leigh-NT1 and Leigh-iPS1 exhibited normal 46XY karyotypes and Leigh-NT2 exhibited a XXXY tetraploid karyotype. d, Leigh-NT1 and Leigh-iPS1cells expressed OCT4 and NANOG. Scale bars, 200 μm. e, Histological analyses of teratoma tumours produced after injections of Leigh-NT1 and Leigh-iPS1 cells into SCID mice. Scale bars, 200 μm. Haematoxylin and eosin staining of teratoma sections identify derivatives of ectoderm, mesoderm and endoderm.
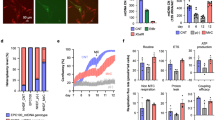
Extended Data Figure 3 Metabolic function in differentiated cells from MELAS iPS cells.
a, OCR/ECAR ratio in MELAS-iPS cells. Mutant MELAS-iPS1 and MELAS-iPS3 displayed significantly decreased OCR/ECAR ratios compared to wild-type MELAS-iPS2 (P < 0.05), indicating a greater reliance on glycolysis (n = 9 per cell line, biological replicates). b, OCR/ECAR ratio in MELAS-iPS cell derived fibroblasts (n = 10 per cell line, biological replicates). c, Immunofluorescence analysis for neural progenitor markers in MELAS-iPS derived NPCs. Scale bar, 100 μm. d, Quantitative analysis of PSC (OCT4 and NANOG) or NPC (SOX1, NESTIN and PAX6) marker expression in MELAS-iPS cells and NPCs (n = 3 per cell line, biological replicates). e, OCR of MELAS-iPS cell derived NPCs (n = 6 per cell line, biological replicates). Error bars are mean ± s.e.m. Significance established with one-way ANOVA with Tukey's multiple comparison test.
Extended Data Figure 4 Metabolic function in differentiated cells from Leigh syndrome PSCs.
a, OCR/ECAR ratio in Leigh-iPS1, Leigh-iPS2 and Leigh-NT1 derived fibroblasts, parental and oocyte donor fibroblasts (n = 9, 8, 10, 9 and 8 per cell line, respectively, biological replicates). b, Immunofluorescence analysis of Leigh-iPS1- and Leigh-NT1-derived skeletal muscle cells labelled with MF20 and myogenin antibodies. Scale bar, 100 μm. c, Cardiomyocyte differentiation efficiency in Leigh-iPS1 and Leigh-NT1 evaluated by FACS for CTnT-Alexa 647 and GATA4-FITC antibodies (n = 3 per cell line, biological replicates). Error bars are mean ± s.e.m. Significance established with one-way ANOVA with Tukey's multiple comparison test.
Extended Data Figure 5 Metabolic function in hESO-NT1 and hESO-8.
a, Immunofluorescence analysis of hESO-NT1 and hESO-8 derived NPCs with nestin and PAX6 antibodies. Scale bar, 100 μm. b, Metabolic profiles of NPCs differentiated from hESO-NT1 and hESO-8 (n = 6 per cell line, biological replicates). c, Immunofluorescence analysis of hESO-NT1 and hESO-8 derived cardiomyocytes with troponin I and NKX2.5 antibodies. Scale bar, 100 μm. d, Efficiency of cardiomyocyte differentiation in hESO-NT1 and hESO-8 evaluated by FACS analysis for CTnT-Alexa 647 and GATA4-FITC antibodies (n = 3 per cell line, biological replicates). e, OCR of hESO-NT1 and hESO-8 derived cardiomyocytes (n = 6 per cell line, biological replicates). Error bars are mean ± s.e.m. Significance established with Student's t-test.
Extended Data Figure 6 RNA-seq analyses of fibroblasts differentiated from MELAS and Leigh syndrome PSCs carrying wild-type and mutant mtDNA.
a, Heat map showing all differentially expressed 1,118 genes (adjusted P value < 0.05) between fibroblasts differentiated from mutant MELAS iPS cells (n = 4 from biological duplicates of MELAS-iPS2 and MELAS-iPS4) and wild-type MELAS iPS cells (n = 4 from biological duplicates of MELAS-iPS1 and MELAS-iPS3). b, Heat map demonstrating differentially expressed 2,950 genes (adjusted P value < 0.05) between fibroblasts derived from wild-type Leigh-NT1 (biological duplicates) and mutant Leigh iPS cells (n = 6 from biological duplicates of Leigh-iPS1, Leigh-iPS2 and Leigh-iPS3). c, Hierarchical clustering using Euclidean distance and average linkage using pvclust, which employs a multiple bootstrap resampling algorithm to calculate the approximately unbiased (AU, red) and bootstrap probability (BU, green) values for cluster distinctions. Hierarchical clustering showed that the Leigh-NT1 fibroblasts were similar to hESO-NT1, hESO-NT2, hESO-7 and hESO-8 fibroblasts. d, Mean log2 normalized counts ± s.e.m. for genes previously reported to be differentially expressed in MELAS cytoplasmic hybrid clones and involved in metabolic and stress response, signalling pathways and epigenetic modifying processes (wild type fibroblast; n = 14 from biological duplicates of 7 independent cell lines; mutant fibroblast n = 10 from biological duplicates of 5 independent cell lines).
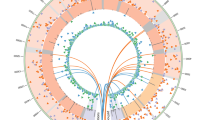
Extended Data Figure 7 RNA‐seq analysis of the mitochondrial transcriptome.
Circular heat map displaying average expression levels for all mitochondrial genes grouped by sample differentiation status and presence or absence of a mutation in the mitochondrial genome (Fib mutant (including primary fibroblasts and PSC derived fibroblasts) with mutant mtDNA n = 14, biological duplicates of 7 independent cell lines; Fib wild type (including primary fibroblasts and PSC derived fibroblasts) with wild‐type mtDNA n = 14, biological duplicates of 7 independent cell lines; PSC mutant (undifferentiated IVF‐ESC, NT‐ESC and iPS cells) with mutant mtDNA n = 3; PSC wild type (undifferentiated IVF‐ESC, NT‐ESC and iPS cells) with wild‐type mtDNA n = 12). The expression of mtDNA‐encoded genes was similar irrespective of 3243A>G or 8993T>G mutations (adjusted P value >0.05).
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3. Table 1 contains a summary of mtDNA variants (SNPs) for MELAS-iPS1, MELAS-iPS2, MELAS-iPS3 and MELAS-fib defined by MiSeq. Table 2 contains a summary of mtDNA variants (SNPs) for Leigh-NT1, Leigh-iPS1 and Leigh-fib defined by MiSeq. Table 3 contains a summary of mtDNA variants (SNPs) for hESO-NT1, hESO-8 and HDF defined by MiSeq (total 12 SNPs differ between hESO-8 and HDF as indicated by red font). (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Ma, H., Folmes, C., Wu, J. et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature 524, 234–238 (2015). https://doi.org/10.1038/nature14546
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14546
This article is cited by
-
iPSC-derived cells lack immune tolerance to autologous NK-cells due to imbalance in ligands for activating and inhibitory NK-cell receptors
Stem Cell Research & Therapy (2023)
-
Induced pluripotent stem cells: ex vivo models for human diseases due to mitochondrial DNA mutations
Journal of Biomedical Science (2023)
-
Molecular basis for maternal inheritance of human mitochondrial DNA
Nature Genetics (2023)
-
Deep exome sequencing identifies enrichment of deleterious mosaic variants in neurodevelopmental disorder genes and mitochondrial tRNA regions in bipolar disorder
Molecular Psychiatry (2023)
-
Chemical reversal of abnormalities in cells carrying mitochondrial DNA mutations
Nature Chemical Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.