Abstract
Cells that reside within a community can cooperate and also compete with each other for resources. It remains unclear how these opposing interactions are resolved at the population level. Here we investigate such an internal conflict within a microbial (Bacillus subtilis) biofilm community: cells in the biofilm periphery not only protect interior cells from external attack but also starve them through nutrient consumption. We discover that this conflict between protection and starvation is resolved through emergence of long-range metabolic co-dependence between peripheral and interior cells. As a result, biofilm growth halts periodically, increasing nutrient availability for the sheltered interior cells. We show that this collective oscillation in biofilm growth benefits the community in the event of a chemical attack. These findings indicate that oscillations support population-level conflict resolution by coordinating competing metabolic demands in space and time, suggesting new strategies to control biofilm growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ben-Jacob, E., Cohen, I. & Levine, H. Cooperative self-organization of microorganisms. Adv. Phys. 49, 395–554 (2000)
Eldar, A. Social conflict drives the evolutionary divergence of quorum sensing. Proc. Natl Acad. Sci. USA 108, 13635–13640 (2011)
Gregor, T., Fujimoto, K., Masaki, N. & Sawai, S. The onset of collective behavior in social amoebae. Science 328, 1021–1025 (2010)
Wingreen, N. S. & Levin, S. A. Cooperation among microorganisms. PLoS Biol. 4, 1486–1488 (2006)
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nature Rev. Microbiol. 8, 15–25 (2010)
Oliveira, N. M., Niehus, R. & Foster, K. R. Evolutionary limits to cooperation in microbial communities. Proc. Natl Acad. Sci. USA 111, 17941–17946 (2014)
Davies, D. Understanding biofilm resistance to antibacterial agents. Nature Rev. Drug Discov. 2, 114–122 (2003)
Donlan, R. M. & Costerton, J. W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 (2002)
Vlamakis, H., Aguilar, C., Losick, R. & Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953 (2008)
Yildiz, F. H. & Visick, K. L. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118 (2009)
Berk, V. et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337, 236–239 (2012)
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999)
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev. Microbiol. 2, 95–108 (2004)
Asally, M. et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl Acad. Sci. USA 109, 18891–18896 (2012)
Wilking, J. N. et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl Acad. Sci. USA 110, 848–852 (2013)
Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R. & Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001)
Gunka, K. & Commichau, F. M. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol. Microbiol. 85, 213–224 (2012)
Stannek, L. et al. Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environ. Microbiol http://dx.doi.org/10.1111/1462-2920.12813 (2015)
Belitsky, B. R. & Sonenshein, A. L. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180, 6298–6305 (1998)
Zeigler, D. R. et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190, 6983–6995 (2008)
Nakano, M. M., Yang, F., Hardin, P. & Zuber, P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J. Bacteriol. 177, 573–579 (1995)
Kleiner, D. Bacterial ammonium transport. FEMS Microbiol. Lett. 32, 87–100 (1985)
Castorph, H. & Kleiner, D. Some properties of a Klebsiella pneumoniae ammonium transport negative mutant (Amt-). Arch. Microbiol. 139, 245–247 (1984)
Boogerd, F. C. et al. AmtB-mediated NH3 transport in prokaryotes must be active and as a consequence regulation of transport by GlnK is mandatory to limit futile cycling of NH4+/NH3. FEBS Lett. 585, 23–28 (2011)
Jayakumar, A., Schulman, I., MacNeil, D. & Barnes, E. M. Jr. Role of the Escherichia coli glnALG operon in regulation of ammonium transport. J. Bacteriol. 166, 281–284 (1986)
Kim, M. et al. Need-based activation of ammonium uptake in Escherichia coli. Mol. Syst. Biol. 8, 616 (2012)
Commichau, F. M., Gunka, K., Landmann, J. J. & Stulke, J. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 190, 3557–3564 (2008)
Detsch, C. & Stulke, J. Ammonium utilization in Bacillus subtilis: transport and regulatory functions of NrgA and NrgB. Microbiology 149, 3289–3297 (2003)
Anyan, M. E. et al. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 111, 18013–18018 (2014)
Irnov, I. & Winkler, W. C. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol. Microbiol. 76, 559–575 (2010)
Acknowledgements
We would like to thank K. Süel, T. Çağatay, R. Wollman, T. Hwa and M. Elowitz for comments during the writing of the manuscript. A.P. is a Simons Foundation Fellow of the Helen Hay Whitney Foundation. J.H. is supported by the UCSD Cell and Molecular Genetics Training Grant. J.G.-O. is supported by the Ministerio de Economia y Competitividad (Spain) and FEDER, under project FIS2012-37655-C02-01, and by the ICREA Academia Programme. This research was funded by the National Institutes of Health, National Institute of General Medical Sciences Grant R01 GM088428 and the National Science Foundation Grant MCB-1450867 (both to G.M.S.).
Author information
Authors and Affiliations
Contributions
G.M.S., J.L., M.A. and J.G.-O. designed the research; J.L. performed the experiments; J.L., J.H. and M.A. performed the data analysis; M.G.-S. and J.G.-O. performed the mathematical modelling; D.D.L., S.L. and M.A. made the bacteria strains; and G.M.S., A.P., J.L., J.H., M.G.-S. and J.G.-O. wrote the manuscript. All authors discussed the manuscript.
Corresponding author
Extended data figures and tables
Extended Data Figure 1 Characterization of biofilm growth oscillations.
a, Top: growth rate over time of an oscillating colony. Bottom: the pressure that drives media flow in the microfluidic chamber is constant over time (see Methods, ‘Microfluidics’ section). b, Top: growth rate of an oscillating colony. Bottom: period of each oscillation cycle, measured peak to peak. The error bars (±20 min) are determined by the imaging frequency (1 frame per 10 min). The period slightly increases over time (see also Extended Data Fig. 6f and Supplementary Information, ‘Mathematical Model’).
Extended Data Figure 2 Roles of carbon and nitrogen in biofilm growth oscillations.
a, Effect of increasing carbon (glycerol) or nitrogen (glutamate) availability on the oscillations. While increasing glutamate by five times of the normal MSgg levels leads to quenching of the oscillation, increasing glycerol by five times does not. b, Colony growth of mutant strain with rocG deletion. Bacillus subtilis NCIB 3610 has two glutamate dehydrogenases (GDH), rocG and gudB. While gudB is constitutively expressed, rocG expression is subject to carbon catabolite repression18. The oscillatory growth of the rocG deletion strain indicates that carbon-source-dependent regulation of rocG expression is not required for biofilm oscillations.
Extended Data Figure 3 Fourier transform of biofilm growth rates before and after perturbations.
The perturbations are: a, addition of 1 mM glutamine; b, addition of 1 mM ammonium; and c, addition of 1 mM IPTG to induce Phyperspank-RocG. The error bars show standard deviations (n = 3 colonies for each condition). The arrows indicate the frequency of oscillations for each condition before perturbation (left) and the lack of oscillations after perturbation (right).
Extended Data Figure 4 Measurements of cell growth within oscillating biofilms.
a, Top: visual representation of the method through which difference movies are generated (Methods, ‘Data analysis’ section). Growth is represented by white pixels, and lack of growth is indicated by black pixels. Film strip (middle) and growth area over time (bottom) of an oscillating colony. Dashed lines show the position of each image on the time trace. Scale bar, 100 µm. b, Top left: schematic of a biofilm. Top right: high magnification phase contrast image of biofilm periphery focused at the bottom layer of cells. Bottom panel: time traces depicting elongation rates of single cells in grey. Highlighted in red is the single cell time trace for the cell outlined in red in the top-right panel. The periodic slowdown of the growth of individual peripheral cells is responsible for the observed periodic reduction in biofilm expansion.
Extended Data Figure 5 Effects of external ammonium on biofilm development.
a, Addition of external ammonium (red shading, 1 mM) represses expression from the PnasA-YFP reporter (black), but does not affect expression from a constitutive reporter (Phyperspank-CFP + 1 mM IPTG, grey). b, Removal of external ammonium (red shading, 13 mM) causes halting of colony growth.
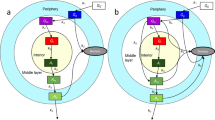
Extended Data Figure 6 Mathematical model of biofilm growth.
a, The model describes the dynamics of two cell populations in a biofilm, interior and peripheral. As the biofilm grows, there is a constant distance between the interior population and the biofilm edge. b–e, Bifurcation diagrams showing systematic analysis on the effects of external glutamine, external glutamate, ammonium uptake, and GDH overexpression, respectively. The red lines correspond to the extrema of oscillations in peripheral glutamate (stable limit cycle). The solid black line denotes stable fixed point. The dashed black line corresponds to an unstable fixed point. The vertical grey lines highlight the state of the system for each nutrient addition experiment shown in Fig. 3. f, Model prediction of oscillation period as function of interior cell fraction in the whole biofilm. g, h, Sensitivity analysis of oscillation period (g) and modulation depth (h) to changes in model parameters. Modulation depth is defined as the amplitude of the oscillations divided by the mean value. Grey colour denotes parameter regions where the system does not oscillate.
Extended Data Figure 7 Temporal profile of cell death within an oscillating biofilm.
Top: colony growth rate. Bottom: average fluorescence intensity of a cell death marker (Sytox green, 1 µM, Life Technologies) from the same colony shown in the top panel.
Extended Data Figure 8 Effect of external attack with hydrogen peroxide (H2O2, 0.15% v/v) or chloramphenicol (CM, 5 µg ml−1).
Top: cell death shown by Sytox green (1 µM). Middle and bottom: colony growth shown by image differencing (see Extended Data Fig. 4a and Methods, ‘Data analysis’). Scale bar, 100 µm. The white dashed lines indicate colony edge.
Extended Data Figure 9 Effect of GDH induction on cell growth.
Wild-type and Phyperspank-RocG (uninduced or induced with 10 mM IPTG) strains were grown in liquid culture (MSgg medium, 30 °C). Cell generation times were measured using OD600. Error bars show standard deviations (n = 3 replicates).
Extended Data Figure 10 Growth rate oscillations persist in various mutant strains.
a, opp operon deletion (deficient in quorum sensing). b, comX deletion (deficient in quorum sensing). c, tapA operon deletion (extracellular matrix component deletion). d, tapA operon overexpression (Phyperspank-tapA operon, 1 mM IPTG). e, hag deletion (deficient in swimming and swarming). These results show that the corresponding genes and processes are not required for biofilm oscillations.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data and Supplementary Tables 1-2. (PDF 952 kb)
Time-lapse video showing the growth of a biofilm.
To better visualize the growth oscillation, the periphery region the biofilm is shown. Also shown is the quantification of growth rate over time. Growth rate is defined as the speed at which the biofilm edge expands along the radial direction. (MP4 1294 kb)
Time-lapse video showing the onset of growth oscillations in a growing biofilm.
The associated time trace shows the growth rate as a function of colony diameter, where oscillatory growth only emerges when the biofilm exceeds a minimum size. (MP4 4171 kb)
Time-lapse video showing in white the region of active expansion for an oscillating biofilm.
The region of expansion is determined by the consecutive image subtraction (difference) approach described in the manuscript (Extended Data Fig. 4a, and Methods: Data Analysis). The associated time trace shows the growth rate as a function of time. (MP4 1078 kb)
Rights and permissions
About this article
Cite this article
Liu, J., Prindle, A., Humphries, J. et al. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554 (2015). https://doi.org/10.1038/nature14660
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14660
This article is cited by
-
Optogenetic spatial patterning of cooperation in yeast populations
Nature Communications (2024)
-
Spatial biology of Ising-like synthetic genetic networks
BMC Biology (2023)
-
Spatial transcriptome uncovers rich coordination of metabolism in E. coli K12 biofilm
Nature Chemical Biology (2023)
-
Pulcherriminic acid modulates iron availability and protects against oxidative stress during microbial interactions
Nature Communications (2023)
-
BMX: Biological modelling and interface exchange
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.