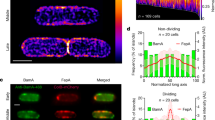
Abstract
Gram-negative bacteria inhabit a broad range of ecological niches. For Escherichia coli, this includes river water as well as humans and animals, where it can be both a commensal and a pathogen1,2,3. Intricate regulatory mechanisms ensure that bacteria have the right complement of β-barrel outer membrane proteins (OMPs) to enable adaptation to a particular habitat4,5. Yet no mechanism is known for replacing OMPs in the outer membrane, an issue that is further confounded by the lack of an energy source and the high stability6 and abundance of OMPs5. Here we uncover the process underpinning OMP turnover in E. coli and show it to be passive and binary in nature, in which old OMPs are displaced to the poles of growing cells as new OMPs take their place. Using fluorescent colicins as OMP-specific probes, in combination with ensemble and single-molecule fluorescence microscopy in vivo and in vitro, as well as molecular dynamics simulations, we established the mechanism for binary OMP partitioning. OMPs clustered to form ∼0.5-μm diameter islands, where their diffusion is restricted by promiscuous interactions with other OMPs. OMP islands were distributed throughout the cell and contained the Bam complex, which catalyses the insertion of OMPs in the outer membrane7,8. However, OMP biogenesis occurred as a gradient that was highest at mid-cell but largely absent at cell poles. The cumulative effect is to push old OMP islands towards the poles of growing cells, leading to a binary distribution when cells divide. Hence, the outer membrane of a Gram-negative bacterium is a spatially and temporally organized structure, and this organization lies at the heart of how OMPs are turned over in the membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
van Elsas, J. D., Semenov, A. V., Costa, R. & Trevors, J. T. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183 (2011)
Tenaillon, O., Skurnik, D., Picard, B. & Denamur, E. The population genetics of commensal Escherichia coli. Nature Rev. Microbiol. 8, 207–217 (2010)
Morabito, S. ed. Pathogenic Escherichia coli: Molecular & cellular microbiology (Caister Academic Press, 2014)
Vogel, J. & Papenfort, K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611 (2006)
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003)
Moon, C. P., Zaccai, N. R., Fleming, P. J., Gessmann, D. & Fleming, K. G. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl Acad. Sci. USA 110, 4285–4290 (2013)
Hagan, C. L., Silhavy, T. J. & Kahne, D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011)
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013)
Kleanthous, C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nature Rev. Microbiol. 8, 843–848 (2010)
Housden, N. G. et al. Intrinsically disordered protein threads through the bacterial outer membrane porin OmpF. Science 340, 1570–1574 (2013)
Shapiro, L., McAdams, H. H. & Losick, R. Why and how bacteria localize proteins. Science 326, 1225–1228 (2009)
Housden, N. G., Loftus, S. R., Moore, G. R., James, R. & Kleanthous, C. Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding. Proc. Natl Acad. Sci. USA 102, 13849–13854 (2005)
Kurisu, G. et al. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nature Struct. Biol. 10, 948–954 (2003)
Buchanan, S. K. et al. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 26, 2594–2604 (2007)
Mullineaux, C. W., Nenninger, A., Ray, N. & Robinson, C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J. Bacteriol. 188, 3442–3448 (2006)
Verhoeven, G. S., Dogterom, M. & den Blaauwen, T. Absence of long-range diffusion of OmpA in E. coli is not caused by its peptidoglycan binding domain. BMC Microbiol. 13, 66 (2013)
Smit, J. & Nikaido, H. Outer membrane of Gram-negative bacteria. XVIII. Electron microscopic studies on porin insertion sites and growth of cell surface of Salmonella typhimurium. J. Bacteriol. 135, 687–702 (1978)
Ursell, T. S., Trepagnier, E. H., Huang, K. C. & Theriot, J. A. Analysis of surface protein expression reveals the growth pattern of the Gram-negative outer membrane. PLOS Comput. Biol. 8, e1002680 (2012)
Browning, D. F. et al. Mutational and topological analysis of the Escherichia coli BamA protein. PLoS ONE 8, e84512 (2013)
Webb, C. T. et al. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 422, 545–555 (2012)
Roder, F. et al. Reconstitution of membrane proteins into polymer-supported membranes for probing diffusion and interactions by single molecule techniques. Anal. Chem. 83, 6792–6799 (2011)
White, J. C., DiGirolamo, P. M., Fu, M. L., Preston, Y. A. & Bradbeer, C. Transport of vitamin B12 in Escherichia coli. Location and properties of the initial B12-binding site. J. Biol. Chem. 248, 3978–3986 (1973)
Casuso, I. et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nature Nanotechnol. 7, 525–529 (2012)
Goose, J. E. & Sansom, M. S. Reduced lateral mobility of lipids and proteins in crowded membranes. PLOS Comput. Biol. 9, e1003033 (2013)
Jarosławski, S., Duquesne, K., Sturgis, J. N. & Scheuring, S. High-resolution architecture of the outer membrane of the Gram-negative bacteria Roseobacter denitrificans. Mol. Microbiol. 74, 1211–1222 (2009)
Stewart, E. J., Madden, R., Paul, G. & Taddei, F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3, e45 (2005)
Laloux, G. & Jacobs-Wagner, C. How do bacteria localize proteins to the cell pole? J. Cell Sci. 127, 11–19 (2014)
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014)
Ieva, R., Tian, P., Peterson, J. H. & Bernstein, H. D. Sequential and spatially restricted interactions of assembly factors with an autotransporter β domain. Proc. Natl Acad. Sci. USA 108, E383–E391 (2011)
Konovalova, A., Perlman, D. H., Cowles, C. E. & Silhavy, T. J. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl Acad. Sci. USA 111, E4350–E4358 (2014)
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006)
Penfold, C. N. et al. Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells. J. Bacteriol. 186, 4520–4527 (2004)
Garinot-Schneider, C., Pommer, A. J., Moore, G. R., Kleanthous, C. & James, R. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 260, 731–742 (1996)
Roder, F., Birkholz, O., Beutel, O., Paterok, D. & Piehler, J. Spatial organization of lipid phases in micropatterned polymer-supported membranes. J. Am. Chem. Soc. 135, 1189–1192 (2013)
Yin, J. et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl Acad. Sci. USA 102, 15815–15820 (2005)
Ray, N., Nenninger, A., Mullineaux, C. W. & Robinson, C. Location and mobility of twin arginine translocase subunits in the Escherichia coli plasma membrane. J. Biol. Chem. 280, 17961–17968 (2005)
Mashanov, G. I. & Molloy, J. E. Automatic detection of single fluorophores in live cells. Biophys. J. 92, 2199–2211 (2007)
Kusumi, A., Sako, Y. & Yamamoto, M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65, 2021–2040 (1993)
Deich, J., Judd, E. M., McAdams, H. H. & Moerner, W. E. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc. Natl Acad. Sci. USA 101, 15921–15926 (2004)
Leake, M. C. et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 (2006)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675 (2012)
Zinchuk, V., Wu, Y., Grossenbacher-Zinchuk, O. & Stefani, E. Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nature Protocols 6, 1554–1567 (2011)
Degrip, W. J., Vanoostrum, J. & Bovee-Geurts, P. H. Selective detergent-extraction from mixed detergent/lipid/protein micelles, using cyclodextrin inclusion compounds: a novel generic approach for the preparation of proteoliposomes. Biochem. J. 330, 667–674 (1998)
Vogelsang, J. et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. 47, 5465–5469 (2008)
Espenel, C. et al. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 182, 765–776 (2008)
Krementsov, D. N. et al. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11, 1401–1414 (2010)
Marrink, S. J., Risselada, H. J., Yefimov, S., Tieleman, D. P. & de Vries, A. H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007)
Bond, P. J., Wee, C. L. & Sansom, M. S. Coarse-grained molecular dynamics simulations of the energetics of helix insertion into a lipid bilayer. Biochemistry 47, 11321–11331 (2008)
Bond, P. J. & Sansom, M. S. Insertion and assembly of membrane proteins via simulation. J. Am. Chem. Soc. 128, 2697–2704 (2006)
Klauda, J. B., Brooks, B. R. & Pastor, R. W. Dynamical motions of lipids and a finite size effect in simulations of bilayers. J. Chem. Phys. 125, 144710 (2006)
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. (2011)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Jones, S. & Thornton, J. M. Principles of protein–protein interactions. Proc. Natl Acad. Sci. USA 93, 13–20 (1996)
Javanainen, M. et al. Anomalous and normal diffusion of proteins and lipids in crowded lipid membranes. Faraday Disc. 161, 397–417 (2013)
Oddershede, L., Dreyer, J. K., Grego, S., Brown, S. & Berg-Sorensen, K. The motion of a single molecule, the lambda-receptor, in the bacterial outer membrane. Biophys. J. 83, 3152–3161 (2002)
Spector, J. et al. Mobility of BtuB and OmpF in the Escherichia coli outer membrane: implications for dynamic formation of a translocon complex. Biophys. J. 99, 3880–3886 (2010)
Leake, M. C. et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl Acad. Sci. USA 105, 15376–15381 (2008)
Jeanteur, D. et al. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl Acad. Sci. USA 91, 10675–10679 (1994)
Acknowledgements
We thank the University of Oxford Micron facility and the University of York Biology Technology Facility for access to microscopy facilities. The authors also thank S. Buchanan and K. Jakes for anti-BamC antibody and ColIa plasmids, I. Henderson for HA-tagged BamA constructs, C. Robinson for TatA–GFP construct, K. Heurlier and J. Pullen for PCR screening of deletion strains, A. Sharma for the design of the inactivating disulfide bond in ColIa, and P. E. Milhier and his group for providing optimised PATRACK single-molecule tracking software. A.L.D. and M.C. thank T. Reddy for discussions and assistance with diffusion calculations. U.S. and C.G.B. thank the University of York Biology Electronic and Mechanical Workshops for custom fabrications, and G. Mashanov for custom image acquisition software. We also thank B. Berks for comments on the manuscript. C.G.B. acknowledges the Royal Society (2004/R1) and the University of York (Research Priming Fund) for financial support used to develop the TIRFM. P.R. acknowledges the late R. Saadia for his unwavering support. M.S.P.S. acknowledges access to the UK supercomputer ARCHER for molecular dynamics simulations. This work was supported by grants to C.K. and C.G.B. (BBSRC LoLa grant BB/G020671/1), J.P. (Deutsche Forschungsgemeinschaft SFB 944) and M.S.P.S. (BBSRC BB/L002558/1, Wellcome Trust WT092970MA).
Author information
Authors and Affiliations
Contributions
P.R., O.B., J.P., M.S.P.S., C.G.B. and C.K. designed the experiments. N.A.C., assisted by C.G.B. and C.T., collected SMT-TIRFM data and C.T., assisted by N.A.C. and P.R., collected FRAP data for colicin-labelled OMPs using wild-type and deletion E. coli strains. P.R. conducted all ensemble TIRFM experiments on OMP islands. U.S. and C.G.B. designed and built the TIRF microscope at York used for all SMT-TIRFM experiments. O.B. and P.R. conducted all PSM experiments and data analysis. S.J.C. and C.G.B. designed and implemented software scripts used for diffusion simulations and bacterial cell image analysis. C.T., P.R., N.A.C., N.G.H. and R.K. purified all the proteins used in the study. C.T., P.R. and N.A.C. labelled colicins with fluorophores. D.M.Q. and T.J.G. constructed plasmids for colicin constructs and established procedures for growth of wild-type and gene deletion E. coli strains. M.C. and A.L.D. conducted the molecular dynamics simulations and subsequent analysis. C.K. drafted the paper with assistance from P.R., C.G.B., M.S.P.S. and J.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Visualizing the different diffusive behaviours of proteins in the inner and outer membranes of the same E. coli cell.
Single E. coli JM83 bacterial cell visualized by laser-scanning confocal fluorescence microscopy at 37 °C. At the outer membrane (top row) BtuB labelled with ColE9TMR showed no fluorescence recovery after photobleaching a region of interest with a 543 nm laser (rectangle). By contrast, recovery of TatA–GFP fluorescence in the inner membrane of the same cell (bottom row) was observed within a few seconds after photobleaching an identical region with 488 nm laser. The unrestricted mobility of TatA–GFP in the inner membrane accounts for its analogue distribution during cell division (main text, Fig. 1b, f). This experiment was done in duplicate (one representative set of images is presented in the figure). Scale bar, 1 μm.
Extended Data Figure 2 SMT-TIRFM defines the mobility of colicin-labelled BtuB and Cir in vivo.
a, Top, Z-projection of fluorescence intensity (increasing from blue to red) of two Cir–ColIaAF488 complexes on separate JM83 cells. Middle, the fluorescence intensity in consecutive images (30 f.p.s.) is displayed for a typical Cir–ColIaAF488 fluorescent spot. The intensity and single-step photobleaching behaviour were consistent with a single membrane complex being tracked. Bottom, MSD was calculated for single Cir–ColIaAF488 complexes (n = 41) that displayed single-step photobleaching behaviour, could be tracked for at least 1.7 s before photobleaching (error is reported as s.e.m.) and were not immobilized on the quartz surface (MSDend of trajectory > 0.004, determined for surface bound ColIaAF488 in the same samples). The MSD value rapidly approached an asymptotic value that was consistent with restricted lateral diffusion. Linear regression of the MSD for the first 4 time delays (0.033–0.13 s) yielded the planar diffusion coefficient (D ≈ 0.019 μm2 s−1). b, Top, Z-projection of fluorescence intensity (increasing from blue to red) of a BtuB–ColE9AF488 complex on a JM83 cell. Middle, the fluorescence intensity in consecutive images (30 f.p.s.) is displayed for a typical fluorescent spot. The intensity and single-step photobleaching behaviour were consistent with a single membrane complex being tracked. Bottom, MSD was calculated for individual BtuB–ColE9AF488 complexes (n = 62) that displayed single-step photobleaching behaviour, could be tracked for at least 1.7 s before photobleaching (error is reported as s.e.m.) and were not immobilized on the quartz surface (MSDend of trajectory > 0.008, determined for surface bound ColE9AF488 in the same samples). The MSD value rapidly approached an asymptotic value that was consistent with restricted lateral diffusion. Linear regression of the MSD for the first five time delays (0.033–0.17 s) yielded the planar diffusion coefficient (D ≈ 0.013 μm2 s−1). See Extended Data Table 1 for all fitted values (from a minimum of four experimental replicates). c, Comparison of experimental MSD for BtuB–ColE9AF488 complexes from b (open circles) with mean output from Monte Carlo simulations (solid blue line) of 2D diffusion (D = 0.14 μm2 s−1, association time of 0.2 s (k−1), and 15% volume occupancy) in a curved, crowded membrane typical of a rod-shaped bacterium, which is illuminated by an evanescent field (penetration depth = 150 nm). Here lateral D was identical to the value observed for monomeric BtuB diffusing in PSMs (Extended Data Fig. 5c). The lower apparent lateral D measured experimentally ( = 0.013 μm2 s−1) indicates the promiscuous PPIs induce temporal corralling. The dashed (blue) lines are the upper and lower 99.75% confidence limits for the mean (n = 5,000 trajectories). See Methods for details of the Monte Carlo simulations. d, MSD plots (±s.e.m.) for all confined trajectories (500 in each case) of BtuB–ColE9TMR complexes diffusing in PSMs, red data points in Fig. 4b (panels 3 and 4), main text. [BtuB] × 1,000 data (open circles) is compared to data for native levels of BtuB (closed circles) in the presence of native levels of OmpF (equivalent to [BtuB] × 1000). Fits to both sets of data yield values for D and confinement diameter, ∼0.02 μm2 s−1 and 0.4 μm, respectively, that are very similar to those observed in vivo.
Extended Data Figure 3 Probing the roles of cell envelope systems in the restricted diffusion of OMPs.
Laser-scanning confocal FRAP microscopy of E. coli cells labelled with ColE9AF488 for BtuB (a) and ColIaAF488 or ColIaTMR for Cir (b) was used in conjunction with gene deletions or chemical treatments to probe the involvement of various cell envelope systems in the restricted mobility of OMPs. The rationale for these experiments was twofold. First, structures or processes within the Gram-negative cell envelope could be responsible for the restricted mobility of OMPs (Extended Data Figs 1 and 2). Second, FRAP would provide a means of detecting the loss of restricted OMP mobility, resulting in the recovery of fluorescence after photobleaching, if these systems were perturbed. Each panel in the figure shows DIC microscopy images of the bacterial cell followed by pre-bleach, bleach (t = 0) and post-bleach (t = 3 min) fluorescent images of the same cell. All microscopy images are 3.03 × 4.17 μm2. Top-to-bottom; CCCP treatment (0.1 mM) of E. coli JM83 cells assessed the impact of dissipating the proton-motive force across the inner membrane. E. coli BZB1107 cells (ompR vlamB ompF::Tn5) are deficient for the major outer membrane porins OmpF and OmpC. E. coli BW25113 rfaC (JW3596) is a deep-rough mutant in which the outer core of the LPS is truncated. E. coli BW25113 lpp (JW1667) is a deletion of Braun’s lipoprotein, an outer membrane lipoprotein that is one of the most abundant proteins in E. coli. ∼40% of Lpp is covalently attached to the underlying peptidoglycan. E. coli BW25113 tonB (JW5195) is deleted for TonB, a protein that spans the periplasm and couples the proton-motive force across the inner membrane with transport of nutrients through outer membrane proteins such as BtuB and Cir. ColIa depends on TonB for import into bacteria. E. coli BW25113 tolA (JW0729) is deleted for TolA, a protein that spans the periplasm and couples proton-motive force with stabilization of the outer membrane. ColE9 depends on TolA for import into bacteria. Other E. coli K-12 deletion strains tested (but not shown) include pal (JW0731), an outer membrane lipoprotein, and ompA (JW0940), both of which have domains that form non-covalent contacts with the peptidoglycan cell wall. As the data in the figure show, no mutation or condition resulted in the recovery of fluorescence in FRAP experiments from which we infer these systems/processes are not responsible for the restricted mobility of OMPs. Finally, we tested a mutation of ColE9 in which the first 83 amino acids of the colicin were deleted (Δ1–83 ColE9) but where the inactivating disulfide across the R-domain remained in place. The N-terminal 83 residues contain protein–protein interaction epitopes for OmpF as well as TolB in the periplasm. No change in FRAP behaviour (not shown) was observed demonstrating that interactions made by the colicin at the cell surface are not responsible for the restricted mobility of the OMP to which it is bound. These experiments were done in duplicate (one representative set of images for each condition is presented in the figure).
Extended Data Figure 4 Estimation of the number of BtuB and Cir proteins within OMP islands.
a, The scatter plots show the distribution of relative intensity of ColE9AF488 and ColIaTMR fluorescence signals when bound to their specific OMPs in bacteria or adhered to surfaces. The right-hand panels show representative TIRFM images that were used for the analysis. Fluorescence signals corresponded to a stack of 100 frames (30 f.p.s.) and were relatively stable in time and space. Scale bars, 1 μm. The mean value (± s.e.m.) for each distribution is shown by the histograms. We normalized the data for colicins adhered to bacteria using the mean intensity for surface-adhered ColE9AF488 and ColIaTMR (300 nM), assuming these correspond to one molecule (∼1 AU). From this normalization, OMP islands on average contained ∼7 labelled OMPs although values ranged from 1 to 19 labelled OMPs. The entire experiment was conducted twice, the replicate showing the same mean value of OMPs per island. b, Co-localization of BamA with Cir. TIRFM images (sum of 100 frames) of E. coli JWD3 cells expressing HA-tagged BamA detected by Alexa488-labelled anti-HA antibody1919. Cir was detected by ColIaTMR labelling before and after a 1-h period of growth. BamA shows significant co-localization (31 ± 7% for 112 OMP islands visualized across 20 cells) with Cir within OMP islands. Owing to weak binding of the anti-HA antibody it was not possible to perform more detailed growth experiments as for BamC. c, Co-localization of BamC with Cir/TIRFM images (sum of 100 frames) for E. coli JM83 cells stained with Alexa488-labelled anti-BamC antibody and ColIaTMR showing co-localization within OMP islands that move to the poles in cells undergoing division. Temporal separation of BamC and the Cir labels by a 1-h period of growth, in which old BamC was first blocked with unlabelled antibody, showed that old Cir was localized primarily at the old poles (red label), whereas new BamC-containing islands appeared in non-polar regions of the cell (green label). The average fluorescence distributions and co-localization histograms (error is reported as s.e.m.) shown in b and c are from 20 cells in each case. *P < 0.1 (Student’s t-test); **P < 0.001 (Mann–Whitney test).
Extended Data Figure 5 Influence of membrane components on diffusion in polymer-supported membranes.
a, Analysis of the relative fluorescence intensity of BtuB–ColE9TMR suggested significant self-association of BtuB, which was time and concentration dependent. The grey area corresponds to the range of intensities where single step photobleaching was observed and which were the molecules tracked in our experiments. b, Distribution of diffusion coefficient when tracking LPSBODIPY incorporated into a polymer-supported bilayer at a ratio of 17:1 ratio LPS:BtuB. High concentrations of BtuB or OmpF, but not of TM-MBP, induced significant trapping of some LPS molecules. c, Distribution of diffusion coefficient when tracking BtuB–ColE9TMR incorporated into a polymer supported bilayer that contains a ratio of 17:1 or 0:1 LPS:BtuB. Addition of LPS did not alter significantly the diffusion behaviour of BtuB in these artificial membranes. d, Distribution of diffusion coefficients when tracking TM-MBPCy5 incorporated into a polymer-supported bilayer. High concentrations of BtuB or OmpF, but not of TM-MBP, induced a slight but significant trapping of some TM-MBP molecules. All experiments were done in duplicate (one set are presented in the figure) and differences in diffusion coefficient determined using a non-parametric Mann–Whitney test.
Extended Data Figure 6 Final snapshots of OmpF (yellow) and BtuB (green) positions for coarse grain molecular dynamics simulations.
a–f, Each patch (∼30 × 30 nm2 for a–e, ∼60 × 60 nm2 for f) contained four BtuB (a); nine BtuB (b); two OmpF trimers and two BtuB monomers (c); four OmpF and five BtuB (d); eight OmpF and eight BtuB (e); and eight OmpF and eight BtuB (f). In each case, the single unit cell is darkened and outlined in blue. Lipids are shown in grey (PE) and red (PG). See Extended Data Fig. 7 and Extended Data Table 2 for further details. g, Propensity for BtuB (green) and OmpF (yellow) residues to be at the protein–protein interface based on simulations of BtuB–OmpF mixtures. A propensity greater than one indicates that a residue occurs more frequently at the interaction interface than on the protein surface. h, BtuB residue propensities for residue types at the interface between two BtuB monomers, based on simulations containing OmpF and BtuB (dark green), or just BtuB (light green). The two propensities are similar. Note that the sampling in terms of homo-interactions of BtuBs in the OmpF–BtuB simulation is less than in the simulation containing only BtuB owing to the starting positions of the proteins in OmpF–BtuB simulations, which favour hetero-interactions between BtuB and OmpF. See Methods for details about the calculation of the propensity values.
Extended Data Figure 7 Residues that mediate BtuB–OmpF and BtuB–BtuB interactions in molecular dynamics simulations.
a, b, Residues that mediate BtuB–OmpF (a) and BtuB–BtuB (b) interactions. The interaction matrix charts the frequency of interaction between any pair of BtuB and OmpF residues, as a proportion of the total number of interactions that occurred, from high proportional frequency (dark green) to low (white). Depicted here is a subset of the entire interaction matrix, showing only the residues which engaged in interactions with the other protein over a threshold value; any BtuB residue which had a proportional interaction frequency of more than 1 × 10−3 with any OmpF residue is shown, and similarly for any OmpF residue. On each side, residues with interaction frequency values above approximately one-third of the maximum value of interaction are highlighted in bold. The bar plots show the proportional interaction frequencies of each single BtuB (side) and OmpF (top) residue, for the subset of residues that are shown in the interaction matrix. See Methods for a full mathematical explanation of the interaction value calculations. Bar plots are coloured according to the bar values, from high proportional interaction frequency (dark red) to low (white). This is consistent with the colour scheme in Fig. 4e. Note the matrix in b is symmetric.
Extended Data Figure 8 Binary OMP partitioning and its relationship to biogenesis and organization of OMP islands.
a, Model depicting the appearance of new OMP islands (green) within growing cells containing old OMP islands (red) in which OMP biogenesis has ceased. The TIRFM images of single cells shown alongside the different stages were taken from cell growth experiments (same experiments as shown in Fig. 2, main text). The model highlights how new OMP islands appear mostly at mid-cell, their creation in conjunction with cell elongation forces old OMP islands towards the poles. The result is binary turnover when the cell divides. The bulk of the old OMPs are retained at the poles of repository cells, which are created at every cell division. It remains unclear why OMP biogenesis ceases in old OMP islands. It is also unclear whether the architecture of the poles (for example, membrane curvature) has a role in retaining old OMPs or whether this is entirely a consequence of continued growth and biogenesis in daughter cells. Nevertheless, the outcome of such a mechanism is that within just two divisions cells appear that do not have any of the original old OMPs. The stochastic nature of the process occasionally results in some old OMP islands not partitioning with the old pole (for example, main text Fig. 2b, panel 3, TIRFM image), which likely explains why the process is not a pure binary mechanism (if it were, values of 100% would be expected in the histogram shown in Fig. 1f, main text). We speculate that the lack of intermixing between OMP islands, which is consistent with the absence of fluorescence recovery in in vivo FRAP experiments (Extended Data Fig. 1), may be due to the very high density of OMPs (particularly porins)25 within islands separated by densely packed LPS. b, Model depicting the organization of an individual OMP island and the movement of an OMP within it. OMP islands contain one or more Bam complexes (see main text Fig. 3). Here, we show a single Bam machine having just deposited an OMP (green circle) in the OM. The new OMP diffuses laterally in the membrane but becomes increasingly restricted due to promiscuous interactions with other OMPs (grey circles), which is consistent the confinement experienced by OMPs in vitro and in vivo (main text Fig. 4b and Extended Data Fig. 2) and with Monte Carlo and molecular dynamics simulations (main text Fig. 4c, d and Extended Data Figs 2c, 6 and 7). Only a fraction of the OMPs presumed to be present within an OMP island are shown.
Supplementary information
Binary distribution of ColE9TMR-labelled BtuB in E. coli JM83 cells expressing TatA-GFP followed over several cell divisions.
Red and green channels correspond, respectively, to BtuB-ColE9TMR in the OM and TatA-GFP in the IM (see main text Figure 1b and Methods for details). The corresponding DIC images and an overlap of green and red channels (merge) are also shown. Images were acquired every 30 min, starting from a single cell. All scale bars, 1μm. Owing to technical issues this video is not viewable in our online player. Please download the video to view it. (MOV 530 kb)
BtuB and Cir co-segregate to the poles during E. coli cell division and are both sequestered in repository cells.
Red and green channels correspond, respectively, to Cir-ColIaTMR and BtuB-ColE9AF488 (see Methods for details). The corresponding DIC images and an overlap of green and red channels (merge) are also shown. Images have been acquired every 10 min from the same single cell. All scale bars, 1 μm. Owing to technical issues this video is not viewable in our online player. Please download the video to view it. (MOV 239 kb)
TIRF microscopy data showing diffusion of single receptor complexes in the outer membrane of E. coli.
Cir and BtuB molecules labelled with ColIaAF488 and ColE9AF488, respectively, were tracked in the OM of JM83 cells (see Extended Data Fig. 2a, b and Methods for details). Video frames 1-23 and 24-91 collected at 30 fps show diffusion of single Cir-ColIaAF488 and BtuB-ColE9AF488 complexes, respectively (video playback at 15 fps), scale bar 2 μm (5x digital magnification applied to video images). (MOV 719 kb)
TIRF microscopy data showing photobleaching of BtuB-ColE9AF488 complexes in the outer membrane of E. coli.
Video (1300 frames) collected at 30 fps shows how high-density labelling gives rise to discrete fluorescent patches (OMP islands) and then to single fluorophore labelled OMP as photobleaching progresses. This video complements Video 3, which was acquired following complete photobleaching of the sample and tracking of single fluorophore-labelled ColE9 molecules arriving subsequently. The end point was identical in both experimental approaches. Scale bar, 1 µm (6x digital magnification applied to video images). (MOV 3234 kb)
Biogenesis of new OMP islands in the central regions of dividing E. coli cells pushes old OMP islands towards the poles.
Red and green channels correspond, respectively, to pre-existing BtuB (labelled with ColE9TMR) and newly synthesised BtuB (labelled with ColE9AF488) after 30 min of growth (see main text Figure 2a and Methods for details). The corresponding DIC images and an overlap of green and red channels (merge) are also shown. t = 0 min image was taken before cell growth and subsequent images were acquired every 30 min from the same cell seen at t = 30 min. All scale bars, 1 μm. Owing to technical issues this video is not viewable in our online player. Please download the video to view it. (MOV 451 kb)
Diffusion of BtuB labelled ColE9TMR in a polymer-supported membrane composed of E. coli lipids.
Red trajectories correspond to single molecule tracking of BtuB-ColE9TMR complexes after incorporation into an artificial membrane at a density similar to that of BtuB in vivo. See main text Figure 4a,b and Methods for details. Video collected at 30 f.p.s. Scale bar, 2 μm. (MOV 8897 kb)
Rights and permissions
About this article
Cite this article
Rassam, P., Copeland, N., Birkholz, O. et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 523, 333–336 (2015). https://doi.org/10.1038/nature14461
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14461
This article is cited by
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Single-molecule dynamics show a transient lipopolysaccharide transport bridge
Nature (2023)
-
Physical properties of the bacterial outer membrane
Nature Reviews Microbiology (2022)
-
Peptidoglycan maturation controls outer membrane protein assembly
Nature (2022)
-
Bacterial envelope built to a peptidoglycan tune
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.