Abstract
The alternative non-homologous end-joining (NHEJ) machinery facilitates several genomic rearrangements, some of which can lead to cellular transformation. This error-prone repair pathway is triggered upon telomere de-protection to promote the formation of deleterious chromosome end-to-end fusions1,2,3. Using next-generation sequencing technology, here we show that repair by alternative NHEJ yields non-TTAGGG nucleotide insertions at fusion breakpoints of dysfunctional telomeres. Investigating the enzymatic activity responsible for the random insertions enabled us to identify polymerase theta (Polθ; encoded by Polq in mice) as a crucial alternative NHEJ factor in mammalian cells. Polq inhibition suppresses alternative NHEJ at dysfunctional telomeres, and hinders chromosomal translocations at non-telomeric loci. In addition, we found that loss of Polq in mice results in increased rates of homology-directed repair, evident by recombination of dysfunctional telomeres and accumulation of RAD51 at double-stranded breaks. Lastly, we show that depletion of Polθ has a synergistic effect on cell survival in the absence of BRCA genes, suggesting that the inhibition of this mutagenic polymerase represents a valid therapeutic avenue for tumours carrying mutations in homology-directed repair genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Data deposits
Sequence has been deposited with the BioProject database under accession PRJNA269507.
References
Capper, R. et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 21, 2495–2508 (2007)
Sfeir, A. & de Lange, T. Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597 (2012)
Rai, R. et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 29, 2598–2610 (2010)
Sfeir, A. Telomeres at a glance. J. Cell Sci. 125, 4173–4178 (2012)
van Steensel, B., Smogorzewska, A. & de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 (1998)
Wang, H. et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 65, 4020–4030 (2005)
Audebert, M., Salles, B., Weinfeld, M. & Calsou, P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 356, 257–265 (2006)
Yan, C. T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007)
Simsek, D. et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 7, e1002080 (2011)
Simsek, D. & Jasin, M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature Struct. Mol. Biol. 17, 410–416 (2010)
Okamoto, K. et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494, 502–505 (2013)
Arana, M. E., Seki, M., Wood, R. D., Rogozin, I. B. & Kunkel, T. A. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008)
Hogg, M., Sauer-Eriksson, A. E. & Johansson, E. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 40, 2611–2622 (2012)
Chan, S. H., Yu, A. M. & McVey, M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005 (2010)
Roerink, S. F., van Schendel, R. & Tijsterman, M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 24, 954–962 (2014)
Koole, W. et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nature Commun. 5, 3216 (2014)
Shima, N. et al. Phenotype-based identification of mouse chromosome instability mutants. Genetics 163, 1031–1040 (2003)
Tang, J. et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature Struct. Mol. Biol. 20, 317–325 (2013)
Wang, M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006)
Truong, L. N. et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. USA 110, 7720–7725 (2013)
Bailey, S. M., Cornforth, M. N., Kurimasa, A., Chen, D. J. & Goodwin, E. H. Strand-specific postreplicative processing of mammalian telomeres. Science 293, 2462–2465 (2001)
Certo, M. T. et al. Tracking genome engineering outcome at individual DNA breakpoints. Nature Methods 8, 671–676 (2011)
Bindra, R. S., Goglia, A. G., Jasin, M. & Powell, S. N. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic Acids Res. 41, e115 (2013)
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008)
Fernandez-Vidal, A. et al. A role for DNA polymerase θ in the timing of DNA replication. Nature Commun. 5, 4285 (2014)
Deng, S. K., Gibb, B., de Almeida, M. J., Greene, E. C. & Symington, L. S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nature Struct. Mol. Biol. 21, 405–412 (2014)
Chen, H., Lisby, M. & Symington, L. S. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 (2013)
Kawamura, K. et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 109, 9–16 (2004)
Higgins, G. S. et al. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 1, 175–184 (2010)
Lemée, F. et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl Acad. Sci. USA 107, 13390–13395 (2010)
Shee, C. et al. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife 2, e01222 (2013)
Leung, J. W. et al. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 10, e1004178 (2014)
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006)
Prasad, R. et al. Human DNA polymerase θ possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 37, 1868–1877 (2009)
Celli, G. B. & de Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nature Cell Biol. 7, 712–718 (2005)
Sfeir, A., Kabir, S., van Overbeek, M., Celli, G. B. & de Lange, T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661 (2010)
Acknowledgements
We thank T. de Lange, R. Greenberg, J. Shay, N. Shima, C. Cazaux and R. Wood for providing key reagents for this study. We are grateful to M. Ji, L. Walton Masters, A. Phillips, A. Pinzaru, F. Yeung, P. Tonzi and J. Wong for technical assistance. We thank S. Kabir and F. Lottersberger for critical reading of the manuscript. This work was supported by a grant from the Breast Cancer Alliance (A.S.), V-foundation (A.S.), Department of Defense Breast Cancer Research Program BC134020 (P.A.M.-G.), Pew-Stewart Scholars Award (A.S.), Pew Scholars Award (E.L.-D.), Novartis Advanced Discovery Institute (E.L.-D.), and a grant from the National Institutes of Health (NIH) AG038677 (E.L.-D.). The A.S. laboratory was supported by start-up funds from the Helen L. and Martin S. Kimmel Center for Stem Cell Biology. The K.M.M. laboratory was supported in part by start-up funds from the University of Texas at Austin and from the Cancer Prevention Research Institute of Texas (CPRIT, R116). K.M.M. is a CPRIT scholar.
Author information
Authors and Affiliations
Contributions
A.S., E.L.-D. and P.A.M.-G. conceived the experimental design. P.A.M.-G. and A.S. performed the experiments and analysed the data. E.L.-D. and N.N. performed telomere-sequencing experiments. F.G. and K.M.M. performed experiments related to Polθ localization at DNA breaks. A.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
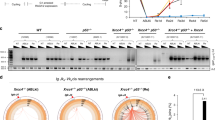
Extended Data Figure 1 Polθ promotes alt-NHEJ repair at dysfunctional telomeres.
(Related to Fig. 1.) a, Immunoblots for TRF1 and RAP1 after 4-OHT-induced depletion of TRF2 from Trf2F/FCre-ERT2 MEFs and co-depletion of TRF1 and TRF2 from Trf1F/F Trf2F/F Ku80−/− Cre-ERT2 cells. Loss of TRF2 is confirmed by the disappearance of RAP1; a TRF2-interacting protein the stability of which depends on TRF2 (refs 35, 36). b, To validate the effect of Polq depletion on alt-NHEJ we monitored the frequency of telomere fusions in shelterin-free Ku80-null cells treated with three independent shPolq vectors. shPolq-1 was used in Fig. 2. Mean values are presented with error bars denoting ± s.e.m. from two independent experiments.
Extended Data Figure 2 Polθ drives chromosomal translocations in mouse cells.
(Related to Fig. 2.) a, Immunobloting for Polθ in MEFs with the indicated genotype and treatment. b, Immunoblot for TRF1 in MEFs with the indicated genotype. Cells were analysed 96 h after Cre induction. c, RAP1 immunoblot (similar to b). d, Western blot analysis for Polθ and Flag–Cas9 in lysates prepared from Polq−/− and Polq+/+ cells after Cas9 expression. Tubulin serves as a loading control. e, Surveyor nuclease assay for Polq−/− and Polq+/+ cells expressing Cas9-gRNA(Rosa26;H3f3b) plasmid. Genomic DNA isolated from cells with the indicated genotype was used as a template to amplify across the cleavage site at either the Rosa26 or the H3f3b locus to assess intra-chromosomal NHEJ. Amplification products were denatured and then re-annealed to form heteroduplexes between unmodified and modified sequences from imprecise NHEJ. The mismatched duplex was selectively cleaved by the Surveyor nuclease at the loops that form at mismatches. f, Signature of translocations in Polq−/− and Polq+/+ cells (see Extended Data Figs 3–4, 5 for complete list of sequences). Table records the total number of translocation events identified following CRISPR-Cas9 induced-cleavage. On average, the same number of nucleotides was deleted at the fusion junction in Polq−/− and Polq+/+ cells. No nucleotide insertions were found in the absence of Polq. Lastly, the percentage of junctions exhibiting microhomology was significantly reduced in cells lacking Polq. g, Scheme depicting Polθ domains. CRISPR/Cas9 gene targeting was used to create two base substitutions at Asp2494Gly and Glu2495Ser, and generate a catalytic-dead polymerase34. h, Sequence analysis of targeted cells. i, Genotyping PCRs of Polq+/+ and PolqCD (catalytically dead allele of Polq) after SacII digestion. j, Immunoblotting to analyse Cas9 expression in Polq+/+ and two independently derived PolqCD clonal cell lines. k, Frequency of chromosomal translocations (der(6)) in Polq+/+ and PolqCD cells. Bars represent mean of four independent experiments ± s.d. (two experiments per clonal cell line). P = 0.006; two-tailed Student’s t-test. PCR products were sequenced to confirm translocation and identify possible insertions.
Extended Data Figure 3 Sequence analysis of translocation junctions in Polq+/+ cells.
(Related to Fig. 2.) Sequences of der(11) breakpoint junction from Polq+/+ cells. Predicted fusion breakpoint based on CRISPR cutting indicated by an arrow. Reference sequence highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing. Nucleotide insertions are marked in red. In cases where insertions extended beyond the sequence included in the lane, the length of the insertion was noted in parenthesis (red). Gaps in the sequence represent nucleotide deletions. The average length of the deletions was noted in Extended Data Fig. 2f. Micro-homology is denoted by blue boxes. Micro-homology embedded in DNA extending beyond the enclosed sequence was noted in parentheses (blue).
Extended Data Figure 4 Sequence analysis of translocation junctions in Polq+/+ cells.
(Related to Fig. 2.) Sequences of der(6) breakpoint junction from Polq+/+ cells. Predicted fusion breakpoint based on CRISPR cutting indicated by an arrow. Reference sequence highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing.
Extended Data Figure 5 Sequence analysis of translocation junctions in Polq−/− cells.
(Related to Fig. 2.) Sequences of der(11) and der(6) breakpoint junction from Polq−/− cells. Predicted fusion breakpoint based on CRISPR cutting indicated by an arrow. Reference sequence is highlighted at the top. The remaining lines represent individual translocations recovered by PCR and subject to Sanger sequencing. It is important to note that insertions were completely lacking at the fusions junctions in Polq−/− cells.
Extended Data Figure 6 Polθ recruitment to DNA breaks.
(Related to Fig. 3.) a, Laser micro-irradiation experiment using HeLa cells expressing Myc–Polθ and treated with ATM inhibitor (KU55933), ATR inhibitor (VE-821) or PARP inhibitor (KU58948). b, Western blot analysis for CHK1 and CHK2 phosphorylation. Cells with the indicated treatment were analysed 2 h after irradiation. c, Immunoblot for PARP1. HeLa cells were treated with PARP1 siRNA and analysed 72 h after siRNA transfection for efficiency of knockdown.
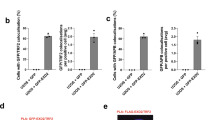
Extended Data Figure 7 PARP1-dependent Polθ recruitment to DNA double-stranded breaks (DSBs).
(Related to Fig. 3.) a, Results from immunofluorescence performed 4 h after induction (1 µM Shield1 ligand, Clontech 631037; 0.5 μM 4-OH tamoxifen) of DSBs by mCherry-LacI-FokI in the U2OS-DSB reporter cells18 transfected with the Myc–Polθ and treated with PARP inhibitor (KU58948). The mCherry signal is used to identify the area of damage and to assess the recruitment of Myc–Polθ to cleaved LacO repeats. b, Table displaying quantification related to a.
Extended Data Figure 8 Polθ suppresses homology-directed repair at dysfunctional telomeres.
(Related to Fig. 3.) a, Western blot analysis for Polθ and LIG3 in shelterin-free Lig4-null MEFs. b, Western blot for TRF1 and RAP1 after 4-OHT treatment of shelterin-free Lig4-deficient cells. c, Metaphase spreads from Trf1F/F Trf2F/F Lig4−/− Cre-ERT2 MEFs, with the indicated shRNA treatment, 96 h after Cre expression. CO-FISH assay was performed using a FITC-OO-(CCCTAA)3 PNA probe (green) and a Tamra-OO-(TTAGGG)3 PNA probe (red). DAPI in blue. Examples of alt-NHEJ-mediated fusion and T-SCE events (HDR) are indicated by white and red arrows, respectively. Examples of T-SCE events reflective of increased HDR in cells treated with shPolq are on the right. d, e, Quantification of telomere fusions by alt-NHEJ in MEFs with the indicated genotype and shRNA treatment. Bars represent mean of two independent experiments ± s.e.m. f, Representative in-gel hybridization to assess 3′ overhang of Trf1F/F Trf2F/F Lig4−/− Cre-ERT2 MEFs with the indicated shRNA treatment after Cre deletion. g, Quantification of the gel in f. The single-stranded DNA/total signal ratios of the ‘+Cre’ samples are expressed relative to the ‘−Cre’ samples for each shRNA treatment. Mean of two independent experiments. h, Graph representing RAD51 accumulation after ionizing radiation treatment of PolqCD, Polq+/+ and Polq−/− embryonic stem cells. Bars represent mean of two independent experiments. P >0.05; two-tailed Student’s t-test.
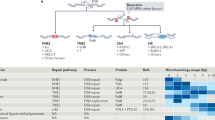
Extended Data Figure 9 Polθ promotes alt-NHEJ and inhibits homology-directed repair at I-SceI-induced DNA breaks.
(Related to Fig. 3.) a, Polθ represses recombination at DSBs induced by I-Sce1. The TLR system was used to measure the relative ratio of end-joining (mCherry) and HDR (enhanced green fluorescent protein (eGFP)) repair of a DSB. A diagram of the TLR is represented. b, The TLR construct was stably integrated into Lig4−/ and Ku80−/− MEFs to avoid the confounding effect of C-NHEJ, and limit end-joining reactions to the alt-NHEJ pathway. Expression of mCherry and eGFP was assessed by flow cytometry 72 h after I-Sce1 and 5′ eGFP donor transduction in cells with the indicated shRNA construct. Percentages of cells are indicated in the plot. c, Quantification of alt-NHEJ and HDR of TLR containing Ku80−/−MEFs after expression of I-Sce1 and 5′ eGFP together with the indicated shRNA construct. Bar graphs represent the mean of three independent experiments ± s.d. P = 0.03; two-tailed Student’s t-test. d, Real-time PCR to monitor the knockdown efficiency of Polq in Ku80−/− and Lig4−/− MEFs. The FACS analysis reported in e and f was carried out without selecting for cells expressing the shRNA-containing plasmid.
Extended Data Figure 10 Polθ is required for survival of recombination-deficient cells.
(Related to Fig. 4.) a, Accumulation of chromosomal aberrancies after Brca1 and Brca2 knockdown in Polq−/− and Polq+/+ MEFs. Quantification of chromosomal aberrancies (chromatid breaks, chromosome breaks and radials) in MEFs stably transduced with lentiviral vectors expressing the indicated shRNA. b, Real-time PCR to confirm the knockdown of Brca1 and Brca2 as in a. c, Quantitative analysis of colony formation in Brca1F/F Cre-ERT2 and Lig4−/− cells after Polq depletion. The number of colonies in control shRNA-treated cells was set to 100%. Mean values are presented with error bars denoting ± s.d. from three independent experiments. d, Real-time PCR to measure the knockdown efficiency of human POLQ in BJ-hTERT, MCF7 and HCC1937 cells and mouse Polq in Brca1F/F Cre-ERT2 cells. e, Quantitative analyses of colony formation in BJ-hTERT, MCF7 and HCC1937 cells after LIG3 inhibition. The number of colonies in control-shRNA-treated cells was set to 100%. The knockdown efficiency for Lig3 was ∼85%. Bars represent mean of two independent experiments ± s.e.m. f. Quantitative analyses of colony formation in PolqCD and Polq+/+ embryonic stem cells after BRCA1 inhibition. The number of colonies in control-shRNA-treated cells was set to 100%. The knockdown efficiency for BRCA1 was >80%. Bars represent mean of two independent experiments ± s.e.m. P = 0.05; two-tailed Student’s t-test.
Supplementary information
Supplementary Information
This file contains Supplementary Data including sequence analysis of telomere fusions using illumina technology and C‐NHEJ junction sequences. (PDF 233 kb)
Source data
Rights and permissions
About this article
Cite this article
Mateos-Gomez, P., Gong, F., Nair, N. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). https://doi.org/10.1038/nature14157
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14157
This article is cited by
-
POLQ inhibition attenuates the stemness and ferroptosis resistance in gastric cancer cells via downregulation of dihydroorotate dehydrogenase
Cell Death & Disease (2024)
-
The ALT pathway generates telomere fusions that can be detected in the blood of cancer patients
Nature Communications (2024)
-
CATI: an efficient gene integration method for rodent and primate embryos by MMEJ suppression
Genome Biology (2023)
-
ZSCAN4 interacts with PARP1 to promote DNA repair in mouse embryonic stem cells
Cell & Bioscience (2023)
-
Clinical and translational advances in ovarian cancer therapy
Nature Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.