Abstract
Evolutionarily conserved SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins form a complex that drives membrane fusion in eukaryotes. The ATPase NSF (N-ethylmaleimide sensitive factor), together with SNAPs (soluble NSF attachment protein), disassembles the SNARE complex into its protein components, making individual SNAREs available for subsequent rounds of fusion. Here we report structures of ATP- and ADP-bound NSF, and the NSF/SNAP/SNARE (20S) supercomplex determined by single-particle electron cryomicroscopy at near-atomic to sub-nanometre resolution without imposing symmetry. Large, potentially force-generating, conformational differences exist between ATP- and ADP-bound NSF. The 20S supercomplex exhibits broken symmetry, transitioning from six-fold symmetry of the NSF ATPase domains to pseudo four-fold symmetry of the SNARE complex. SNAPs interact with the SNARE complex with an opposite structural twist, suggesting an unwinding mechanism. The interfaces between NSF, SNAPs, and SNAREs exhibit characteristic electrostatic patterns, suggesting how one NSF/SNAP species can act on many different SNARE complexes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
3D cryo-EM density maps of ATP- and ADP-bound NSF, and the 20S supercomplex have been deposited in the Electron Microscopy Data Bank (EMDB), with accession codes EMD-6204 (NSF-ATP), EMD-6205 (NSF-ADP), EMD-6206 (20S-state I), EMD-6207 (20S-state II), EMD-6208 (20S-state IIIa), EMD-6209 (20S-state IIIb) and EMD-6210 (V7–20S). The coordinates of atomic models of ATP- and ADP-bound NSF, the 20S supercomplex have been deposited in the Protein Data Bank (PDB) under the accession codes, 3J94, 3J95, 3J96, 3J97, 3J98 and 3J99.
References
Wickner, W. & Schekman, R. Membrane fusion. Nature Struct. Mol. Biol. 15, 658–664 (2008)
Südhof, T. C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013)
Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395, 347–353 (1998)
Weber, T. et al. SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 (1998)
Jahn, R. & Scheller, R. H. SNAREs–engines for membrane fusion. Nature Rev. Mol. Cell Biol. 7, 631–643 (2006)
Söllner, T., Bennett, M. K., Whiteheart, S. W., Scheller, R. H. & Rothman, J. E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993)
Block, M. R., Glick, B. S., Wilcox, C. A., Wieland, F. T. & Rothman, J. E. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc. Natl Acad. Sci. USA 85, 7852–7856 (1988)
Malhotra, V., Orci, L., Glick, B. S., Block, M. R. & Rothman, J. E. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell 54, 221–227 (1988)
Erzberger, J. P. & Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006)
Matveeva, E. A., He, P. & Whiteheart, S. N-Ethylmaleimide-sensitive fusion protein contains high and low affinity ATP-binding sites that are functionally distinct. J. Biol. Chem. 272, 26413–26418 (1997)
Whiteheart, S. W., Schraw, T. & Matveeva, E. N-ethylmaleimide sensitive factor (NSF) structure and function. Int. Rev. Cytol. 207, 71–112 (2001)
Ernst, J. A. & Brunger, A. T. High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J. Biol. Chem. 278, 8630–8636 (2003)
Stein, A., Weber, G., Wahl, M. C. & Jahn, R. Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528 (2009)
Strop, P., Kaiser, S. E., Vrljic, M. & Brunger, A. T. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J. Biol. Chem. 283, 1113–1119 (2008)
Antonin, W., Fasshauer, D., Becker, S., Jahn, R. & Schneider, T. R. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nature Struct. Biol. 9, 107–111 (2002)
Zwilling, D. et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 26, 9–18 (2007)
Rice, L. M. & Brunger, A. Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol. Cell 4, 85–95 (1999)
Bitto, E. et al. Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family. Proteins 70, 93–104 (2008)
Yu, R. C., Hanson, P. I., Jahn, R. & Brunger, A. T. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nature Struct. Biol. 5, 803–811 (1998)
Lenzen, C. U., Steinmann, D., Whiteheart, S. & Weis, W. Crystal structure of the hexamerization domain of N-ethylmaleimide–sensitive fusion protein. Cell 94, 525–536 (1998)
Yu, R. C., Jahn, R. & Brunger, A. NSF N-terminal domain crystal structure: models of NSF function. Mol. Cell 4, 97–107 (1999)
May, A. P., Misura, K. M., Whiteheart, S. W. & Weis, W. I. Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein. Nature Cell Biol. 1, 175–182 (1999)
Furst, J., Sutton, R. B., Chen, J., Brunger, A. T. & Grigorieff, N. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 Å resolution. EMBO J. 22, 4365–4374 (2003)
Chang, L.-F. et al. Structural characterization of full-length NSF and 20S particles. Nature Struct. Mol. Biol. 19, 268–275 (2012)
Moeller, A. et al. Nucleotide-dependent conformational changes in the N-ethylmaleimide sensitive factor (NSF) and their potential role in SNARE complex disassembly. J. Struct. Biol. 177, 335–343 (2012)
Hanson, P. I., Roth, R., Morisaki, H., Jahn, R. & Heuser, J. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
DeLaBarre, B. & Brunger, A. T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nature Struct. Biol. 10, 856–863 (2003)
Huyton, T. et al. The crystal structure of murine p97/VCP at 3.6 Å. J. Struct. Biol. 144, 337–348 (2003)
Davies, J. M., Brunger, A. T. & Weis, W. I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16, 715–726 (2008)
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nature Methods 9, 853–854 (2012)
Hanson, P. I. & Whiteheart, S. W. AAA+ proteins: have engine, will work. Nature Rev. Mol. Cell Biol. 6, 519–529 (2005)
Zhao, C., Matveeva, E. A., Ren, Q. & Whiteheart, S. W. Dissecting the N-ethylmaleimide-sensitive factor: required elements of the N and D1 domains. J. Biol. Chem. 285, 761–772 (2010)
Barnard, R. J., Morgan, A. & Burgoyne, R. D. Stimulation of NSF ATPase activity by alpha-SNAP is required for SNARE complex disassembly and exocytosis. J. Cell Biol. 139, 875–883 (1997)
Cipriano, D. J. et al. Processive ATP-driven substrate disassembly by the N-ethylmaleimide-sensitive factor (NSF) molecular machine. J. Biol. Chem. 288, 23436–23445 (2013)
Winter, U., Chen, X. & Fasshauer, D. A conserved membrane attachment site in alpha-SNAP facilitates N-ethylmaleimide-sensitive factor (NSF)-driven SNARE complex disassembly. J. Biol. Chem. 284, 31817–31826 (2009)
Weber, T. SNAREpins are functionally resistant to disruption by NSF and αSNAP. J. Cell Biol. 149, 1063–1072 (2000)
Zorman, S. et al. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife e03348 (2014)
Vivona, S. et al. Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between α-soluble NSF attachment protein (SNAP) and SNARE complex. J. Biol. Chem. 288, 24984–24991 (2013)
Fasshauer, D., Sutton, R. B., Brunger, A. T. & Jahn, R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA 95, 15781–15786 (1998)
Marz, K. E., Lauer, J. M. & Hanson, P. I. Defining the SNARE complex binding surface of α-SNAP: implications for SNARE complex disassembly. J. Biol. Chem. 278, 27000–27008 (2003)
Clary, D. O., Griff, I. C. & Rothman, J. E. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell 61, 709–721 (1990)
Wilson, D. W. et al. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature 339, 355–359 (1989)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Fasshauer, D., Eliason, W. K., Brunger, A. T. & Jahn, R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry 37, 10354–10362 (1998)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Li, X., Zheng, S. Q., Egami, K., Agard, D. A. & Cheng, Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. J. Struct. Biol. 184, 251–260 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Shaikh, T. R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nature Protocols 3, 1941–1974 (2008)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Scheres, S. H. W., Núñez-Ramírez, R., Sorzano, C. O. S., Carazo, J. M. & Marabini, R. Image processing for electron microscopy single-particle analysis using XMIPP. Nature Protocols 3, 977–990 (2008)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014)
Zhang, X., Shaw, A., Bates, P. & Newman, R. Structure of the AAA ATPase p97. Mol. Cell 6, 1473–1484 (2000)
Penczek, P., Ban, N., Grassucci, R. A., Agrawal, R. K. & Frank, J. Haloarcula marismortui 50S subunit-complementarity of electron microscopy and X-Ray crystallographic information. J. Struct. Biol. 128, 44–50 (1999)
Saad, A. et al. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J. Struct. Biol. 133, 32–42 (2001)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Acknowledgements
We thank W.I. Weis for discussions and reading of the manuscript; S. Kenrick from Wyatt Technology Corporation for technical help with CG-MALS analysis. This research was supported in part by the National Institutes of Health (R37MH63105 to A.T.B., and a subcontract from Grant #5-U01AI082051-05 to A.T.B.; R01GM082893, R01GM098672 and P50GM082250 to Y.C.).
Author information
Authors and Affiliations
Contributions
M.Z., Q.Z., and D.J.C. expressed and purified all protein samples; M.Z. and S.W. collected cryo-EM data; S.W. processed cryo-EM data; M.Z. built and refined the atomic models based on cryo-EM density maps; M.Z. performed the disassembly assay; S.V. performed the CG-MALS experiment; M.Z., S.W., Y.C., and A.T.B. designed experiments, analysed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
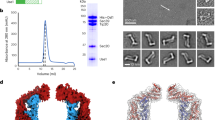
Extended Data Figure 1 Purification of recombinant NSF bound to specific nucleotide and 20S supercomplex.
a, Schematic diagram of the purification steps of NSF. Chromatography columns and buffer conditions are provided. b, Size-exclusion chromatograms corresponding to the coloured steps in panel a. Major peaks are labelled. c, A scheme showing the purification steps of the 20S supercomplex. d, Size-exclusion chromatogram of the 20S supercomplex. Major peaks are labelled. e, SDS–PAGE gel of fractions collected in panel d. The samples were not boiled. f, SDS–PAGE gel of the same fractions as in panel e. The samples were boiled. g, SDS–PAGE-based SNARE disassembly assay of the truncated neuronal SNARE complex. This complex is stable in SDS without boiling. Disassembly by NSF/αSNAP is observed as a function of time.
Extended Data Figure 2 3D reconstruction of ATP-bound NSF by single-particle cryo-EM.
a, A representative electron micrograph (out of 1,150 micrographs) of ATP-bound NSF particles in vitreous ice. b, Selected 2D class averages (6 out of 50). c, Plots of the angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown, with either the z axis or the y axis pointing out towards the viewer. Two corresponding re-projection images of the final density map are shown under the plots. d, Selected slice views of the final reconstruction. The slice numbers are indicated. e, FSC curves for the 3D density map after RELION post-processing. The resolution is estimated to be 4.2 Å by the gold-standard refinement criterion, as indicated by the red arrow. The FSC curve between the refined atomic model and the 3D density map is shown in blue. f, FSC curves for cross-validation. Black, model versus summed map (full data set); green, model versus half map 1 (used for test refinement); orange, model versus half map 2 (not used for test refinement). See Methods for details. g, 3D density map coloured according to the local resolution as estimated by ResMap.
Extended Data Figure 3 Representative densities from the cryo-EM reconstructions of ATP-bound NSF and the 20S supercomplex.
a–g, ATP-bound NSF. Density maps were sharpened by XMIPP using a B-factor of −123 Å2. a, Representative densities (black mesh, 7.8σ) for an α-helix and a β-strand of the D1 domain with the refined model (coloured sticks) superposed. b, Representative density (black mesh, 7.0σ) of the β-sheet of the α/β subdomain of D1 (chain C) with the refined model (yellow Cα ribbon) superposed. c, Density (black mesh, 7.0σ) and model (yellow Cα ribbon) for the D1 and D2 domains of chain B. Note that the linker between the two domains is well resolved. d, De novo model (yellow cartoon) of the D1 domain built from the cryo-EM density map (black mesh, 7.0σ). The arrangement of the subdomains and nucleotide is illustrated in the inset. The pore loop (YVG motif) and two α-helices: α2 from the α/β subdomain, and α7 from the α subdomain are highlighted in the red dotted boxes. e, Density (black mesh, 6.5σ) and model (yellow Cα ribbon) of the ATP binding pocket of the D1 domain (chain C). Motifs that are typical for AAA+ ATPases are indicated. f, Superposition of the crystal structure of the ATP-bound D2 domain with Mg2+(ref. 19, PDB accession code: 1NSF, coloured sticks and balls), and the cryo-EM density map (black mesh, 7.6σ) of ATP-bound NSF (density of chain C). The crystal structure was docked into the density as a rigid body without any refinement. Note that no Mg2+ was present in the samples for cryo-EM studies, but the ATP molecule and the protein coordinates from the crystal structure match the cryo-EM density well. g, Nucleotide-binding sites of the D1 domains from ATP-bound NSF. The density (translucent surface, chains A–E: 8.2σ, chain F: 5.0σ) of each D1 domain is shown together with the built model in ribbon representation. The nucleotide-binding pockets are highlighted by dotted circles. Five out of the six D1 domains show clear density for ATP. h–l, Representative densities (translucent surface, 4.7σ) from the reconstruction for state I of the 20S supercomplex with the model (cartoon) superposed. All densities are representative except for the N domain in panel j, which represents the better-resolved half of the N domain densities (12 out of 24 cases).
Extended Data Figure 4 3D reconstruction of ADP-bound NSF by single-particle cryo-EM.
a, A representative electron micrograph (out of 840 micrographs) of ADP-bound NSF particles in vitreous ice. b, Selected 2D class averages (6 out of 30). c, Selected focused 2D class averages (5 out of 10). The first image shows the focused classification mask, which locates the flipped N domains. d, Plots of angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown, with either the z axis or the y axis pointing out towards the viewer. Two corresponding re-projection images of the final density map are shown under the plots. e, Selected slice views of the final reconstruction. Slice numbers are indicated. f, FSC curve for the 3D density map after RELION post-processing. The resolution is estimated to be 7.6 Å by the gold-standard refinement criterion. g, 3D density map coloured according to the local resolution as estimated by ResMap.
Extended Data Figure 5 Detrimental effect of imposing C6 symmetry on the reconstruction of ADP-bound NSF and C3 symmetry on the reconstruction of the 20S supercomplex.
a, For the NSF maps, in order to visualize densities of the N domains, an unsharpened map is displayed (translucent surface, C1: 1.2σ, C6: 0.6σ) together with the sharpened map using no symmetry (C1) or C6 symmetry during reconstruction (coloured surface, C1: 5.9σ, C6: 7.0σ). For the reconstruction that uses C6 symmetry, symmetric densities for the N domains at top and side positions appear in the unsharpened map, however, these densities cannot be matched to the crystal structure of the N domain. Likewise, the D1 domains appear compressed and cannot be fit well using the structure of the D1 domain that we obtained by asymmetric reconstruction. b, Reconstruction of state I of the 20S supercomplex without symmetry (C1) or with C3 symmetry. Density maps are shown in coloured surfaces similar to Fig. 3 (C1: 4.7σ, C3: 4.9σ). The C3 averaging causes the D1 domains to display alternating up and down positions. The density for the SNARE complex is a featureless rod without the characteristic left-handed twist of the four α-helix bundle. Densities for only three SNAPs emerge, but without any interpretable features (for example, there are no grooves between helices), preventing a match with the crystal structure of the known homologue of αSNAP, Sec17. The N domain densities are weak and none of them exhibit the expected kidney shape.
Extended Data Figure 6 Comparison of AAA+ ATPase domains from ATP- and ADP-bound NSF structures.
a, Unrolling of the ATPase domains of ATP-bound NSF. Two orthogonal views are shown. Individual chains are aligned based on the D2 domains (white) to show the split-washer arrangement of the D1 domains. b, Unrolling of the ATPase domains of ADP-bound NSF. Individual chains are aligned as in panel a. Dotted boxes in panels a and b highlight the α2 helices of the D1 domains in order to help with visualization of the relative positions. The six protomer chains are rainbow coloured as in Fig. 2. c, Superposition of the six D1 domains of ATP-bound NSF based on the α/β subdomains (white). The relative positions of α7 helices from the α subdomains are illustrated in the inset. d, Corresponding superposition of the ADP-bound NSF D1 domains. e, Superposition of the five D1 domains (without chain F) of ATP-bound NSF (grey), and six D1 domains of ADP-bound NSF (white) based on the α/β subdomains. The α7 helices from the α subdomains are highlighted by red dotted boxes. The relative translation of the α7 helices between the ATP-bound state and the ADP-bound state is shown in the inset.
Extended Data Figure 7 Comparison of ATP- and ADP-bound NSF structures, and ATPase domains of ATP-bound NSF and 20S supercomplex.
a–c, Surface representations of the D2, D1 and N domains of ATP- and ADP-bound NSF (looking down from the N-terminal side of the NSF hexamer). The maximum diameters of the D2 and D1 rings, and the interface areas (calculated by PISA66) between ATPase domains are indicated. Each protomer chain is coloured as in Fig. 2. The D1 ring is also shown in panel c and coloured white to help with visualization. d–f, The ATPase domains of the structure of the 20S supercomplex (state I) were superposed on the ATP-bound NSF using the D1 ring as the reference for the fit. Six protomer chains from ATP-bound NSF are rainbow coloured as in Fig. 2. The ATPase domains of the 20S supercomplex are colored in white and grey. Note that the density of chain F in the reconstruction of ATP-bound NSF alone is poorly resolved (Fig. 1b), whereas in the 20S reconstruction it is well defined, although the overall resolution of the 20S reconstruction is lower. d, Side views. e, Top view of the D2 rings. Each individual D2 domain is labelled. Percentages of interface area change (from NSF to 20S) between the D2 domains are provided in the figure. The interface areas between the D2 domains are similar in the NSF and 20S structures, except for a significant increase (12%) between chains D and E for 20S compared to NSF alone. f, Top view of the D1 rings. Each D1 domain is labelled, with the split between chains A and F indicated by a black arrow. The translation of the α7 helix in α subdomain of chain A is illustrated in the inset. Percentages of interface area change (from NSF to 20S) between the D1 domains are shown. Three interfaces stay the same; the one between chains A and B decreases, whereas those between chains E and F, and chains F and A increase significantly.
Extended Data Figure 8 3D reconstruction of 20S supercomplex by single-particle cryo-EM.
a, A representative electron micrograph (out of 610 micrographs) of the 20S supercomplex in its original purification buffer recorded using the TF20 microscope and the TVIPS TemCam-F816 CMOS camera. The inset shows selected 2D class averages (5 out of 50). b, A representative electron micrograph (out of 2,459 micrographs) of 20S supercomplex in the buffer containing additional 0.05% Nonidet P-40 recorded using the TF30 Polara microscope and the Gatan K2 Summit detector. c–g, Results from this imaging condition. c, Selected 2D class averages (6 out of 50). d, Plots of angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown, with either the z axis or the y axis pointing out towards the viewer. Two corresponding re-projection images of the final density map are shown under the plots. e, Selected slice views of the final reconstruction. Slice numbers are indicated. Slices from different layers are framed in different colours: SNAREs and αSNAPs: yellow, N domains: pink, D1 ring: blue, and D2 ring: purple. f, FSC curves for the 3D density maps of the four states after RELION post-processing. The estimated resolution ranges from 7.6 Å to 8.4 Å as estimated by the gold-standard refinement criterion. g, 3D density map coloured using local resolution estimated by ResMap. The right panel shows a cut-through view of the interior of the map. c–e and g are results from a subclass representing state I.
Extended Data Figure 9 Purification and 3D reconstruction of V7-20S supercomplex.
a, Size-exclusion chromatogram of the V7-20S supercomplex. Major peaks are labelled. Only fraction 10 was concentrated and used for single-particle cryo-EM. b, SDS–PAGE gel of fractions collected in panel a. The samples were not boiled. c, SDS–PAGE gel of the same fractions as in panel b. The samples were boiled. d, A representative electron micrograph (out of 993 micrographs) of the V7-20S supercomplex. e, Selected 2D class averages (6 out of 50). f, FSC curve for the 3D density map after RELION post-processing. The estimated resolution is 8.0 Å as estimated by the gold-standard refinement criterion.
Extended Data Figure 10 CG-MALS characterization of αSNAP–SNARE subcomplex.
a, Concentration gradient setup for the experiment that measures the binding between αSNAP and truncated neuronal SNARE complex. b, Measured molar mass for different components. Note that there were two independent runs for αSNAP over the specified concentration ranges. c, Measured molecular mass of αSNAP–SNARE (truncated) subcomplex converted from light scattering over the concentration gradient. The experimental data are represented by blue dots. Simulated curves with different αSNAP:SNARE (truncated) stoichiometry are shown. The best fit is 4:1. d, Measured molecular mass of the αSNAP-V7-SNARE subcomplex calculated from light scattering over the concentration gradient. The experimental data are represented by blue dots. Simulated curves with different αSNAP:V7-SNARE stoichiometry are shown. The best fit is 2:1. e, Calculated mole fractions of different αSNAP–SNARE (truncated) species over the concentration gradient based on 4:1 stoichiometry. f, Calculated mole fractions of different αSNAP–V7-SNARE species over the concentration gradient based on 2:1 stoichiometry.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and Supplementary References. (PDF 374 kb)
Cryo-EM map and structure of ATP-bound NSF
In the first 20 seconds, the unsharpened map is displayed. Its contour is varied to show the densities of the flexible N domains. The color scheme of the atomic model is identical to that of Figure 1. From the 21st second to the end of the video, the sharpened map is displayed. The atomic model is colored the same as in Figure 2. (MOV 29469 kb)
Conformational changes starting from ATP-bound NSF and transitioning to ADP-bound NSF
For clarity, shown are only the two N domains that are flipped along the sides of the ATPase rings. The color scheme is identical to that of Figure 1. Top and bottom views are displayed on the left, and two side views are displayed on the right. Notice that the D1 subunits near the lower edge of the “split washer” rotate outwards, facilitating the flip of the corresponding N domains. The trajectory was computed using the “morph” utility in UCSF Chimera by interpolation between the structures of the ATP-bound state (starting point) and the ADP-bound state (end point) of NSF. (MOV 1000 kb)
Sharpened map of the 20S supercomplex (state I)
The color scheme is identical to that of Figure 3. (MOV 15311 kb)
Tightening of the D1 ATPase ring upon binding of αSNAPs and SNARE complex
For clarity, the D2 ATPase ring is colored in grey. The D1 domains are colored using the same scheme as in Fig. 2. Both a top view and a side view are shown. The trajectory was computed using the “morph” utility in UCSF Chimera by interpolation between the structures of ATP-bound NSF (starting point) and State I of the 20S supercomplex (end point). (MOV 5585 kb)
Movement of the spire relative to the ATPase rings between States IIIa and IIIβ
For clarity, the base (ATPase rings) is colored in grey, and the spire (αSNAP-SNARE subcomplex) and N domains of NSF are colored using the same scheme as in Figure 4. Both a side view and a top view are shown. The trajectory was computed using the “morph” utility in UCSF Chimera by interpolation between the structures of states IIIa (starting point) and IIIb (end point) of the 20S supercomplex. (MOV 3027 kb)
Rights and permissions
About this article
Cite this article
Zhao, M., Wu, S., Zhou, Q. et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61–67 (2015). https://doi.org/10.1038/nature14148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14148
This article is cited by
-
Mechanisms of SNARE proteins in membrane fusion
Nature Reviews Molecular Cell Biology (2024)
-
Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy
Molecular Biomedicine (2022)
-
Identification of two pathways mediating protein targeting from ER to lipid droplets
Nature Cell Biology (2022)
-
Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative
Nature Communications (2021)
-
Extreme parsimony in ATP consumption by 20S complexes in the global disassembly of single SNARE complexes
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.