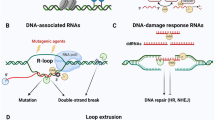
Abstract
The vast majority of the mammalian genome has the potential to express noncoding RNA (ncRNA). The 11-subunit RNA exosome complex is the main source of cellular 3′–5′ exoribonucleolytic activity and potentially regulates the mammalian noncoding transcriptome1. Here we generated a mouse model in which the essential subunit Exosc3 of the RNA exosome complex can be conditionally deleted. Exosc3-deficient B cells lack the ability to undergo normal levels of class switch recombination and somatic hypermutation, two mutagenic DNA processes used to generate antibody diversity via the B-cell mutator protein activation-induced cytidine deaminase (AID)2,3. The transcriptome of Exosc3-deficient B cells has revealed the presence of many novel RNA exosome substrate ncRNAs. RNA exosome substrate RNAs include xTSS-RNAs, transcription start site (TSS)-associated antisense transcripts that can exceed 500 base pairs in length and are transcribed divergently from cognate coding gene transcripts. xTSS-RNAs are most strongly expressed at genes that accumulate AID-mediated somatic mutations and/or are frequent translocation partners of DNA double-strand breaks generated at Igh in B cells4,5. Strikingly, translocations near TSSs or within gene bodies occur over regions of RNA exosome substrate ncRNA expression. These RNA exosome-regulated, antisense-transcribed regions of the B-cell genome recruit AID and accumulate single-strand DNA structures containing RNA–DNA hybrids. We propose that RNA exosome regulation of ncRNA recruits AID to single-strand DNA-forming sites of antisense and divergent transcription in the B-cell genome, thereby creating a link between ncRNA transcription and overall maintenance of B-cell genomic integrity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Sequence Read Archive
Data deposits
All the RNA-seq data sets have been deposited in the Sequence Read Archive under accession number SRP042355 and in the BioProject database under accession number PRJNA248775.
References
Schneider, C. & Tollervey, D. Threading the barrel of the RNA exosome. Trends Biochem. Sci. 38, 485–493 (2013)
Alt, F. W., Zhang, Y., Meng, F. L., Guo, C. & Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152, 417–429 (2013)
Keim, C., Kazadi, D., Rothschild, G. & Basu, U. Regulation of AID, the B-cell genome mutator. Genes Dev. 27, 1–17 (2013)
Klein, I. A. et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 (2011)
Chiarle, R. et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011)
Storb, U. Why does somatic hypermutation by AID require transcription of its target genes? Adv. Immunol. 122, 253–277 (2014)
Pavri, R. et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143, 122–133 (2010)
Basu, U. et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 (2011)
Sun, J. et al. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes Dev. 27, 1821–1833 (2013)
Andrulis, E. D. et al. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420, 837–841 (2002)
Richard, P. & Manley, J. L. Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 (2009)
Economides, A. N. et al. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc. Natl Acad. Sci. USA 110, E3179–E3188 (2013)
Flynn, R. A., Almada, A. E., Zamudio, J. R. & Sharp, P. A. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl Acad. Sci. USA 108, 10460–10465 (2011)
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B. & Sharp, P. A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013)
Wu, X. & Sharp, P. A. Divergent transcription: a driving force for new gene origination? Cell 155, 990–996 (2013)
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008)
Andersen, P. R. et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nature Struct. Mol. Biol. 20, 1367–1376 (2013)
Andersen, P. K., Jensen, T. H. & Lykke-Andersen, S. Making ends meet: coordination between RNA 3′-end processing and transcription initiation. Wiley Interdiscip. Rev. RNA 4, 233–246 (2013)
Flynn, R. A. & Chang, H. Y. Active chromatin and noncoding RNAs: an intimate relationship. Curr. Opin. Genet. Dev. 22, 172–178 (2012)
Seila, A. C. et al. Divergent transcription from active promoters. Science 322, 1849–1851 (2008)
Allmang, C. et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410 (1999)
Ntini, E. et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature Struct. Mol. Biol. 20, 923–928 (2013)
Liu, M. et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451, 841–845 (2008)
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature Genet. 43, 830–837 (2011)
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA 109, 3879–3884 (2012)
Rhee, H. S. & Pugh, B. F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012)
Schulz, D. et al. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155, 1075–1087 (2013)
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 (2012)
Castellano-Pozo, M. et al. R-loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 52, 583–590 (2013)
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chedin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012)
Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 20, 123–128 (1998)
Frendewey, D. et al. The loss-of-allele assay for ES cell screening and mouse genotyping. Methods Enzymol. 476, 295–307 (2010)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010)
Jolly, C. J., Klix, N. & Neuberger, M. S. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 25, 1913–1919 (1997)
Oliveira, T. Y. et al. Translocation capture sequencing: a method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods 375, 176–181 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010)
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013)
Acknowledgements
We thank L. Symington, S. Goff, S. Ghosh, S. Silverstein, C. Lima, F. Chédin, L. Macdonald, C.-S. Lin and O. Couronne for critical input and reagents. This work was supported by grants from the National Institutes of Health (NIH; 1DP2OD008651-01) and the National Institute of Allergy and Infectious Diseases (1R01AI099195-01A1) (to U.B.); NIH (1R01CA185486-01; 1R01CA179044-01A1; 1U54CA121852-05) (to R.R.).
Author information
Authors and Affiliations
Contributions
E.P. and U.B. planned studies; E.P., J.W., G.R., R.R. and U.B. interpreted data. Experiments were performed as follows: E.P. and J.C., mouse model generation, CSR and SHM; E.P., RNA-seq; G.R. and J. L., ChIP, DRIP and CRISPR/Cas9; J.W., bioinformatic studies; A.N.E. advised on the mouse model construct; R.R. oversaw bioinformatics; E.P. and U.B. wrote the manuscript, which was further refined by all the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Exosc3 gene targeting and functional validation of the Exosc3COIN allele.
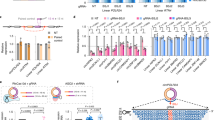
a, Schematic of the Exosc3COINneo BAC targeting vector. Blue shaded boxes indicate Exosc3 exons 1–4. Lox sites are represented by triangles. The GFP-expressing gene trapping module is represented by green arrows. Upstream, downstream and internal homology arms are 41.3, 81.5 and 2.4 kb, respectively. b, Confirmation of Exosc3COINneo/+ targeted embryonic stem (ES)-cell clone AF8. The loss-of-allele (LOA) assay32 was used to screen ES-cell clones for wild-type allele copy number at defined locations within Exosc3 introns 1 and 3 that have been modified to allow for distinction between wild-type and COINneo alleles by TaqMan-based qPCR. A probe for a non-targeted locus, Wnt2b, served as an internal qPCR standard for both copy number and total input DNA. Data represent mean values from six technical replicates. Error bars represent s.d. Ct, cycle threshold. c, HindIII restriction map of the wild-type (WT), COIN and COINinv alleles of Exosc3. The black shaded box indicates the location of the probe used for Southern blotting in Fig 1b. d, Flow cytometric analysis of GFP expression in naive or 4-OHT-treated, LPS plus IL-4-stimulated B-cell cultures. Indicated Exosc3 genotypes are on a ROSA26CreERt2/+ background. One pair of littermate mice was used. Three biological replicates were performed. e, Profile of RNA-seq mapped reads at the Exosc3 locus from 4-OHT-treated, LPS plus IL-4-stimulated B-cell cultures. Indicated Exosc3 genotypes are on a ROSA26CreERt2/+ background. Four biological replicates were performed. f, Immunoblot analysis of Exosc3 and AID protein expression in whole cell extracts from 4-OHT-treated, LPS plus IL-4 stimulated B-cell cultures. Actin was used as a loading control. Wild type, Exosc3WT/WT ROSA26CreERt2/+; C/C, Exosc3COIN/COIN ROSA26CreERt2/+. One pair of littermate mice was used. Two technical replicates were performed. g, Exosc3COIN:Exosc3COINinv ratio in germinal centre B cells determined by qPCR copy number analysis (three technical replicates, error bars represent s.d.).
Extended Data Figure 2 Exosc3-deficient B cells are impaired in CSR and SHM.
a, Representative flow cytometric analysis for surface IgG1 on purified B cells treated with 4-OHT, and stimulated with LPS plus IL-4. Numbers indicate the percentage of GFP+ B220+ B cells having isotype switched to IgG1. One pair of littermate mice was used. Three biological replicates were performed. b, Quantitative RT–PCR time-course analysis of Aicda mRNA expression in naive (day 0) or 4-OHT-treated (days 2–4), LPS plus IL-4 stimulated B-cell cultures. Indicated Exosc3 genotypes are on a ROSA26CreERt2/+ background. Expression levels are normalized to cyclophilin (Ppia) and plotted relative to naive Exosc3COIN/+. Six littermate pairs of each genotype were used. Data represent mean values from three technical replicates. Error bars represent s.d. **P < 0.01 (t-test). c, Quantitative RT–PCR analysis of Ighg1 switch region intron expression. Primers were designed to amplify a region of the Ighg1 GLT intron upstream of the Ighg1 switch region core repeat, but downstream of the Ighg1 non-coding I exon. Two independent pairs of littermate mice of each genotype were used to obtain total RNA from B-cell cultures treated with 4-OHT and stimulated with LPS plus IL-4. Indicated genotypes are on a ROSA26CreERt2/+ background. Data represent mean values from three technical replicates. Error bars represent s.d. Two biological replicates were performed. d, Schematic of the targeted AicdaCre allele. An open reading frame comprising a nuclear localization signal fused to Cre recombinase was used to disrupt the ATG start codon in exon 1 of Aicda. Exons are represented as numbered boxes. e, Specific induction of AicdaCre activity upon LPS plus IL-4 stimulation of B cells. Flow cytometric analysis of AicdaCre activity (as determined by GFP expression) in B220+ and B220− naive splenocyte populations (top panel). AicdaCre induction in LPS plus IL-4 stimulated B-cell cultures (bottom panel). One pair of littermate mice was used. f, AicdaCre is a functional null allele. CSR to IgG1 isotype is abrogated in AicdaCre/Cre homozygous B cells stimulated with LPS plus IL-4. Numbers above gate indicate the percentage of GFP+ B cells having isotype switched to IgG1. One pair of littermate mice was used. g, SHM analysis of Peyer’s patch derived GFP+ germinal centre B cells. Mutation frequencies were determined by sequencing a 645 bp intronic region downstream of the JH4 gene segment of the immunoglobulin heavy chain (IgH) locus. Two littermate pairs of each genotype were used. Two biological replicates were performed. Mutation frequencies represent mean values. P values were determined by proportion test. h, Flow cytometric analysis of Peyer’s patch derived germinal centre B cells from Exosc3COIN/+ and Exosc3COIN/COIN mice on a Cd19Cre/+ background were identified as B220+ PNAhi populations. The percentage of germinal centre B cells amongst all B220+ cells is indicated. One pair of littermate mice was used. Three biological replicates were performed.
Extended Data Figure 3 Proliferation analysis of Exosc3-deficient B cells.
a, FACS analysis indicating the percentage of GFP-negative (left gate) and GFP-positive (right gate) B cells 3.5 days after LPS stimulation. One pair of littermate mice was used. Two biological replicates were performed. b, Kinetic analysis of GFP-positive B-cell accumulation at indicated time points post-LPS stimulation. Indicated Exosc3 genotypes are on a AicdaCre/+ background. One pair of littermate mice was used. Two biological replicates were performed. c, Proliferation analysis determined by VPD450 dilution at 1.5 and 2.5 days post-LPS stimulation. One pair of littermate mice was used. d, Quantitative RT–PCR analysis of Exosc3, Aicda and GFP mRNA expression in GFP+ cells at 3.5 days post-LPS stimulation. Expression levels are normalized to β-actin and plotted relative to Exosc3COIN/+. One pair of littermate mice was used. Data represent mean values from three technical replicates. Error bars represent s.d.
Extended Data Figure 4 Transcriptome analysis of Exosc3-deficient B cells.
a, Genome-wide expression level analysis upstream and downstream of TSS region for expressed protein coding genes. Coding genes with FPKM >1 were determined to be expressed. Analysis was restricted to coding genes that do not have any known genes within a 4 kb upstream boundary. Indicated genotypes are on a ROSA26CreERt2/+ background. One sex-matched littermate pair was used. Two biological replicates were performed. b, Replicate analysis of genome-wide studies. Plots indicate the expression levels of individual genes in Exosc3WT/WT and Exosc3COIN/COIN B cells treated with 4-OHT and stimulated with LPS plus IL-4 from two separate littermate pairs. B cells were purified, cultured and FACS sorted, and RNA was purified and sequenced by RNA-seq all independently between the two experiments. Indicated genotypes are on a ROSA26CreERt2/+ background. Pearson correlation is indicated. c, The distribution of observed lengths for all gapped xTSS-RNAs in Exosc3-deficient B cells. Data were compiled from two biological replicates. d, Scatter plot indicating weak correlation between expression of downstream coding transcript and upstream gapped xTSS-RNA at divergently transcribed loci in Exosc3-deficient B cells. Pearson correlation is indicated. e–g, Profile of RNA-seq mapped reads at the β-actin locus (e) (Actb; 7.6 kb window), Il2rg locus (f) (5.4 kb window) and Ung locus (g) (12 kb window). Indicated genotypes are on a ROSA26CreERt2/+ background and B-cell cultures were treated with 4-OHT and stimulated with LPS plus IL-4. Four biological replicates were performed.
Extended Data Figure 5 xTSS-RNA expression at AID target genes.
a, Quantification of xTSS-RNA expression levels of AID target genes via quantitative RT–PCR from two independent experiments. Indicated genotypes are on a ROSA26CreERt2/+ background. b, Northern blot analysis of xTSS-RNA expression at Myc and Igsγ1 loci. WT, Exosc3WT/WT ROSA26CreERt2/+; C/C, Exosc3COIN/COIN ROSA26CreERt2/+. c–e, Profiles of RNA-seq mapped reads at the Pax5 (c) (72 kb window displaying exons 1–6), Cd83 (d) (22 kb window), and Myc (e) (9 kb window) loci. Red arrows highlight the presence of xTSS-RNA. Four biological replicates were performed. f, Quantification of xTSS-RNA expression levels for AID target genes Myc, Igg1, Cd83, Pim1, Pax5 and Cd79b was obtained from RNA-seq RPKM values from two independent experiments. Indicated genotypes are on a ROSA26CreERt2/+ background. **P < 0.01 (t-test).
Extended Data Figure 6 Translocation hotspots are enriched for xTSS-RNA expression.
a, Strand-specific RNA-seq mapped reads at AID target genes Myc, Cd83, Pim1, Pax5 and Cd79b. Green and red peaks indicate sense and antisense reads, respectively. Red bars represent RefSeq annotation of gene exons. Asterisks indicate the location of TSSs. Arrows indicate the orientation of coding strand transcript. Data were compiled from two biological replicates. b, Boxplot analysis of the level of expression of xTSS-RNAs at various genes reported to undergo recurrent AID-dependent translocations at DNA double-strand breaks generated within the Igh (left panel) or Myc (right panel) loci. Boxplots represent median values compiled from two biological replicates. Whiskers represent 99% of data values. **P < 0.01 (Wilcoxon rank-sum test). c, The list of 40 genes that show an overlap of translocation hotspots and xTSS-RNA expression (from Fig. 3c) was evaluated directly for xTSS-RNA levels (left panel) and mRNA levels (right panel). Statistical analysis was as described in b. **P < 0.01; NS, not significant (Wilcoxon rank-sum test). d, xTSS-RNA expression levels in Exosc3-deficient B cells at non-recurrent and recurrent AID-dependent translocation sites in the B-cell genome. Data were compiled from two biological replicates. **P < 0.01 (Wilcoxon rank-sum test). e, Statistical analysis of the probability of identification of 40 random xTSS-RNA-expressing genes solely based on expression level. Ten-thousand control group genes were randomly selected that were expressed at similar levels as translocation hotspots genes. Specifically, to generate one random control group, we exhausted all translocation hotspots to find genes with similar expression levels (difference of RPKM < 0.5), and randomly picked up one for each hotspot. Ten-thousand gene lists were obtained that contain 88 genes and share the same expression profile with the translocation hotspots list. We then simulate the distribution of genes containing xTSS-RNA by overlapping the random control groups and actual xTSS-RNA gene list. The binomial fitting (red curve) shows that the number of overlapping genes of real translocation hotspots is significantly higher than random controls. **P < 0.01 (binomial distribution).
Extended Data Figure 7 RNA exosome substrate antisense transcripts are expressed within gene bodies and regulatory regions containing AID-induced translocations.
a, Association of genes with TSS-RNA expression, antisense transcripts and AID-induced translocations. The xTSS-RNA and antisense transcripts groups were compiled from four and two biological replicates, respectively. b, Examples of genes with asRNA transcription (Exosc3WT/WT in blue and Exosc3COIN/COIN in red) at regions that have been shown to have translocations from the Igh locus (translocations indicated in black). Data were compiled from two biological replicates. c, Translocations present in the upstream regulatory regions of AID target gene Cd83 (top panel) occur over regions of RNA exosome-sensitive antisense transcription. Data were compiled from two biological replicates.
Extended Data Figure 8 Genome-wide analysis of xTSS-RNA expression at genes with AID, Spt5 and RPA occupancy.
a, Scatter plot of antisense (left) and sense RNA transcription (right) in Exosc3WT/WT and Exosc3COIN/COIN transcriptomes. Data were compiled from two biological replicates. b, Genome-wide analysis of xTSS-RNA expression at genes that are expressed and possess or lack AID and/or Spt5 occupancy. Values beneath each group represent the number of genes with indicated occupancy. P values were determined by Wilcoxon rank-sum test. c, Overlap of genes with xTSS-RNA transcription (pink), recurrent AID-dependent chromosomal translocations (green) and RPA occupancy in the mouse B-cell genome (brown). The xTSS-RNA group was compiled from four biological replicates. **P < 0.01 (Fisher’s exact test).
Extended Data Figure 9 RNA exosome and AID recruitment to divergently transcribed promoter regions.
a, ChIP was performed using anti-H3S10ph or control IgG. Quantitative PCR and data analysis were performed as described in Fig. 4c. **P < 0.01 (t-test). b–d, Representative plots of highly expressed non-divergent genes used as controls for ChIP experiments in Fig. 4. These genes are Cmas (b), Fam107b (c) and March2 (d). Four biological replicates were performed. e, Exosc3 occupancy at divergent and non-divergent promoters. ChIP was performed using anti-Exosc3 (Genway) or control rabbit IgG. Quantitative PCR was performed using primers specific for sequences upstream of the indicated gene TSS. Data are represented as Exosc3 enrichment relative to input. Data represent mean values from three technical replicates. Error bars represent s.d. **P < 0.01 (t-test). f, g, CRISPR/Cas9-mediated deletion strategy of Cd83 (f) and Pim1 (g) xTSS-RNA-expressing regions in CH12F3 B cells. Locations of CRISPR/Cas9 guide RNAs (red markings), genotyping primers (blue triangles), ChIP primers (green triangles), and numbered exons (blue boxes) are indicated. h, Genotyping of Cd83 and Pim1 xTSS-RNA region-deleted CH12F3 clones. i, Cd83 and Pim1 mRNA expression in xTSS-RNA region-deleted CH12F3 cells. Data represent mean values from three technical replicates. *P < 0.05 (t-test). j, Deletion of Cd83 xTSS-RNA-expressing region impairs SHM. Parental CH12F3 or Cd83 xTSS-RNA region-deleted CH12F3 cells were transduced with lentiviral AID and mutation frequency was determined within a 488 bp region beginning approximately 150 bp downstream of the Cd83 TSS. All mutations were derived from unique clonal amplified sequences. Impairment of Cd83 SHM in Cd83 xTSS-RNA region-deleted cells is disproportionally greater than mRNA expression change observed in i. Number of sequenced clones for parental and Cd83 xTSS-RNA region-deleted CH12F3 was 69 and 102, respectively. Background mutation frequency was determined using uninfected control CH12F3 cells and subtracted from the mutation frequencies indicated. **P < 0.01 (proportion test).
Extended Data Figure 10 A model of RNA exosome recruitment to divergently transcribed promoters or at DNA sequences that promote RNA Pol II stalling.
a, Divergent transcription of mRNA in the sense direction recruits RNA exosome and AID following stalling due to various transcription impediments (G-richness in IgH switch sequences is one example). Transcription stalling leading to RNA exosome recruitment occurs more often on the antisense strand due to formation of short asRNAs27. Similarly, in the body of transcribed genes, stalled RNA Pol II generates asRNA transcripts, leading to RNA exosome and AID recruitment. b, Stalled transcripts either close to the TSS or within the body of genes generate DNA–RNA hybrids. These DNA–RNA hybrids contain RPA-coated ssDNA structures that are targets of AID.
Supplementary information
Supplementary Figure 1
Overlap of AID translocation hotpots in 44 genes containing break points with IgH (shown in black bars) with antisense RNA (asRNA) expression from Exosc3COIN/COIN (pink) and Exosc3WT/WT (blue) B cells. (PDF 2040 kb)
Supplementary Table 1
A list of all the annotated transcript units, with mm9 coordinates, strand specificity, and category, that were used for transcriptome analysis. (XLSX 1966 kb)
Supplementary Table 2
A spreadsheet of all the annotated mRNAs and subfamilies of non-coding RNAs (mm9 coordinates listed) investigated in this study to analyze the transcriptome of Exosc3-deficient B cells. RPKM values are listed from two independent experiments. (XLSX 4159 kb)
Supplementary Table 3
A spreadsheet of all genes that express xTSS-RNAs in Exosc3-deficient B cells. (XLSX 248 kb)
Supplementary Table 4
A spreadsheet of all genes that express xTSS-RNA and are AID-mediated translocation hotspots. (XLSX 41 kb)
Supplementary Table 5
A spreadsheet of 84 genes containing similar xTSS-RNA expression as xTSS-RNA-expressing translocation hotspots. (XLSX 55 kb)
Rights and permissions
About this article
Cite this article
Pefanis, E., Wang, J., Rothschild, G. et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014). https://doi.org/10.1038/nature13580
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13580
This article is cited by
-
RNA processing mechanisms contribute to genome organization and stability in B cells
Oncogene (2024)
-
Classifying gastric cancer using FLORA reveals clinically relevant molecular subtypes and highlights LINC01614 as a biomarker for patient prognosis
Oncogene (2021)
-
Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells
Nature Genetics (2021)
-
Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery
Nature Reviews Molecular Cell Biology (2020)
-
Splicing, genome stability and disease: splice like your genome depends on it!
Current Genetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.