Abstract
The balance between stem cell self-renewal and differentiation is controlled by intrinsic factors and niche signals1,2. In the Drosophila melanogaster ovary, some intrinsic factors promote germline stem cell (GSC) self-renewal, whereas others stimulate differentiation3. However, it remains poorly understood how the balance between self-renewal and differentiation is controlled. Here we use D. melanogaster ovarian GSCs to demonstrate that the differentiation factor Bam controls the functional switch of the COP9 complex from self-renewal to differentiation via protein competition. The COP9 complex is composed of eight Csn subunits, Csn1–8, and removes Nedd8 modifications from target proteins4,5. Genetic results indicated that the COP9 complex is required intrinsically for GSC self-renewal, whereas other Csn proteins, with the exception of Csn4, were also required for GSC progeny differentiation. Bam-mediated Csn4 sequestration from the COP9 complex via protein competition inactivated the self-renewing function of COP9 and allowed other Csn proteins to promote GSC differentiation. Therefore, this study reveals a protein-competition-based mechanism for controlling the balance between stem cell self-renewal and differentiation. Because numerous self-renewal factors are ubiquitously expressed throughout the stem cell lineage in various systems, protein competition may function as an important mechanism for controlling the self-renewal-to-differentiation switch.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Morrison, S. J. & Spradling, A. C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008)
Li, L. & Xie, T. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21, 605–631 (2005)
Xie, T. Control of germline stem cell self-renewal and differentiation in the Drosophila ovary: concerted actions of niche signals and intrinsic factors. Wiley Interdiscip. Rev. Dev. Biol. 2, 261–273 (2013)
Wei, N. et al. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8, 919–924 (1998)
Cope, G. A. & Deshaies, R. J. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 (2003)
Lin, H. & Spradling, A. C. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol. 159, 140–152 (1993)
Xie, T. & Spradling, A. C. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251–260 (1998)
Xie, T. & Spradling, A. C. A niche maintaining germ line stem cells in the Drosophila ovary. Science 290, 328–330 (2000)
McKearin, D. & Ohlstein, B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121, 2937–2947 (1995)
Ohlstein, B. & McKearin, D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124, 3651–3662 (1997)
Shen, R., Weng, C., Yu, J. & Xie, T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl Acad. Sci. USA 106, 11623–11628 (2009)
Li, Y., Minor, N. T., Park, J. K., McKearin, D. M. & Maines, J. Z. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl Acad. Sci. USA 106, 9304–9309 (2009)
Chen, D. & McKearin, D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 13, 1786–1791 (2003)
Song, X. et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 131, 1353–1364 (2004)
Chen, D. & McKearin, D. M. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159–1170 (2003)
Lin, H., Yue, L. & Spradling, A. C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947–956 (1994)
Venken, K. J., He, Y., Hoskins, R. A. & Bellen, H. J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006)
Oron, E. et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129, 4399–4409 (2002)
Song, X., Zhu, C. H., Doan, C. & Xie, T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296, 1855–1857 (2002)
Doronkin, S., Djagaeva, I. & Beckendorf, S. K. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev. Cell 4, 699–710 (2003)
Ou, C. Y., Lin, Y. F., Chen, Y. J. & Chien, C. T. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16, 2403–2414 (2002)
Djagaeva, I. & Doronkin, S. Dual regulation of dendritic morphogenesis in Drosophila by the COP9 signalosome. PLoS ONE 4, e7577 (2009)
Schwechheimer, C. et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382 (2001)
Ni, J. Q. et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature Methods 8, 405–407 (2011)
Hofmann, K. & Bucher, P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem. Sci. 23, 204–205 (1998)
Chia, N. Y. et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320 (2010)
Venken, K. J. T. et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nature Methods 6, 431–434 (2009)
Yu, J. et al. MTMR4 attenuates transforming growth factor β (TGFβ) signaling by dephosphorylating R-Smads in endosomes. J. Biol. Chem. 285, 8454–8462 (2010)
Acknowledgements
We thank Y. Yamashita, the Developmental Studies Hybridoma Bank and the Bloomington Drosophila Stock Center for reagents, Xie laboratory members for discussions, and D. Chao and R. Krumlauf for critical comments on the manuscript. This work was supported by the Stowers Institute for Medical Research (T.X.), the National Institutes of Health (GM64428, T.X.), the National Natural Science Foundation of China (31370909, L.P.) and the Ministry of Science and Technology of China (2012CB518900, L.P.).
Author information
Authors and Affiliations
Contributions
L.P., S.W., T.L. and T.X. conceived the project. L.P., S.W., T.L., C.W., X.S., J.K.P., J.Y., H.T., D.M.M. and T.X. collected and analysed the data. J.S., Z.-H.Y., D.A.C. and J.N. contributed the reagents. L.P., S.W., T.L. and T.X. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Bam interacts with Csn4 in yeast cells.
a, Csn4 contains a PCI/PINT protein interaction domain in its C terminus. The C-terminal 121 amino acid region containing the PCI/PINT domain interacts with Bam in yeast cells. b, c, In a yeast two-hybrid assay, different Bam truncations fused to the Gal4 DNA-binding domain were individually tested for their interaction with the fusion protein between Csn4 and the Gal4 transcriptional activation domain by activating expression of the HIS3 gene, which allows yeast cells to grow on medium lacking histidine. “No Bam” means that only the Gal4 binding domain is used in the assay. The results from b are summarized in c. Red-filled boxes indicate positive interactions, whereas unfilled boxes denote no interaction. d, Co-immunoprecipitation (co-IP) experiments show that Myc-tagged Csn4 interacted with different truncated Bam proteins tagged by Flag in S2 cells.
Extended Data Figure 2 BAC transgenes for Flag-tagged Csn4 and Csn5.
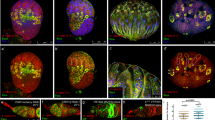
a, The BAC clone contains the 21-kilobase genomic region in which Csn4 is flanked by three genes at the 5′ end and one gene at the 3′ end. A Flag-tag sequence (green symbol) is inserted before the stop codon (Csn4–Flag). b, The BAC clone contains the 21-kilobase genomic region in which Csn5 is flanked by one gene at the 5′ end and four genes at the 3′ end. A Flag-tag sequence (green symbol) is inserted after the translation initiation codon, ATG (Flag–Csn5).
Extended Data Figure 3 Mutations in Csn4 suppress, but mutations in Csn5 and Nedd8 enhance, the bam differentiation defect.
The germarium is indicated by “g”. a, A bamZ/bamΔ86 mutant ovariole contains a tumorous germarium followed by a normal egg chamber (ec) and a tumorous egg chamber (arrow) (the same as in Fig. 1e). b, b′ bamZ/bamΔ86 mutant ovarioles that are also heterozygous for Csn4 contain tumorous germaria and three or more normal egg chambers. c, d, bamZ/bamΔ86 mutant ovarioles that are also heterozygous for Csn5 (c) or Nedd8 (d) contain tumorous germaria and tumorous egg chambers (arrows).
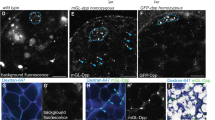
Extended Data Figure 4 Germline-specific knock down of Csn1b, Csn2, Csn6, Csn7 and Csn8 in the adult ovary leads to GSC loss.
a, The combined FLP-Out and Gal4–UAS system for knocking down gene function in adult GSCs and their progeny. An FRT–STOP (transcription stop sequence)–FRT cassette is inserted between the germ-cell-specific nos promoter and the GAL4 coding region to prevent Gal4 expression in germ cells. With heat-shock treatments, FLP expression removes the STOP cassette in most GSCs, leading to Gal4 expression and then short hairpin RNA expression. b–f, Germ-cell-specific Csn1b (c), Csn2 (d), Csn6 (e) and Csn7 (f) germaria contained one (arrow in c, d and f) or no (e) GSCs in contrast to the GFP knock-down germarium, which carried three GSCs (b, arrows). g, The results from the independent knock-down experiment show that germline-specific knock down of Csn1b, Csn2, Csn3, Csn6, Csn7 and Nedd8 caused significant GSC loss in comparison with the GFP knock-down negative control. ***, P < 0.001.
Extended Data Figure 5 Marked Csn4 and Csn5 mutant GSCs are not prone to apoptosis but proliferate more slowly than marked control GSCs.
Marked GSCs and cysts are indicated by dashed circles and arrows, respectively. Unmarked GSCs are highlighted by circles. a, The marked control GSC and cysts are negative for TUNEL labelling. b, b′, The marked Csn4k (b) and Csn4n (b′) mutant GSCs and cysts are negative for TUNEL labelling. c, The marked Csn5n mutant GSCs are negative for TUNEL labelling. Arrowheads indicate dying somatic cells. d, Quantification of TUNEL-positive marked GSCs and cysts. e, f, Marked mutant Csn4k or Csn4n mutant GSCs show significantly lower BrdU incorporation rates than marked control GSCs. Quantification is shown in f (**, P < 0.01; *, P < 0.05). g, h, In contrast to the marked control GSCs (g), the marked Csn4 and Csn5 mutant GSCs have low p-H3 labelling rates (g′). Quantification is shown in h.
Extended Data Figure 6 Inactivation of Csn functions in GSCs does not affect Bmp signalling and E-cadherin expression.
a, a′, Marked (dashed circle) and unmarked (solid circle) control GSCs express p-Mad at similar levels. b, b′, Marked Csn4n mutant (dashed circle) and unmarked control (solid circle) GSCs express p-Mad at similar levels. c, Quantification results show that marked Csn4k and Csn4n mutant GSCs express similar p-Mad levels to marked control GSCs. d–g, p-Mad is expressed normally in Csn1bGSKD (d), Csn2GSKD (e), Csn3GSKD (f) and Csn7GSKD (g) GSCs (dashed circles). h, h′, bam–GFP is repressed in LacZ-negative Csn4n mutant (dashed circle) and LacZ-positive unmarked control (solid circle) GSCs. i–m, LacZ-negative Csn4k (j), Csn4n (k) and Csn5n (l) mutant GSCs (dashed circles) express similar levels of E-cadherin in the stem-cell-niche junction to unmarked control GSCs (solid circles, j–l), as well as marked and unmarked control GSCs (dashed and solid circles, i). Quantification results are shown in m.
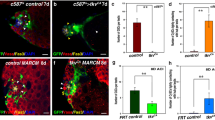
Extended Data Figure 7 Csn4 interferes with the ability of Bgcn to antagonize Bam function.
a, b, In a yeast two-hybrid assay, different Bam truncations fused to the Gal4 DNA-binding domain were individually tested for their ability to interact with the fusion protein between Bgcn and the Gal4 transcriptional activation domain by activating expression of the HIS3 gene, which allows yeast cells to grow on medium lacking histidine. “No Bam” means that only the Gal4 binding domain is used in the assay. The results from a are summarized in b. Red boxes indicate positive interactions; pink boxes represent weak interactions; and unfilled boxes denote no interaction. c, Co-IP experiments show that the central domain of Bam interacts with Bgcn in S2 cells. The asterisk indicates a non-specific protein band. d, d′, Yeast two-hybrid plate (d) and liquid (d′) galactosidase assay results show that Bam interacted significantly more strongly with Csn4 than with Bgcn. e, The C-terminal deletion of the Bam-interacting domain (the C-terminal 121 amino acids) of Csn4 protein destroys Csn4’s ability to compete with Bgcn for binding to Bam. f, A bamΔ86 homozygous germarium contains two GSCs (circle); single germ cells are identified by the spherical spectrosome (arrowhead) and two-cell pairs by the elongated fusome (arrow). These two-cell pairs undergo complete cytokinesis to generate two cystoblasts. g, GSC quantification results for the experiments in Fig. 3d show that germline-specific expression of Csn4 did not change the GSC number in either wild-type or bam heterozygous ovaries. h, Quantification results for Fig. 3e show that Csn4 overexpression suppressed ectopic Bam-expression-induced GSC loss.
Extended Data Figure 8 Csn proteins other than Csn4 promote cystoblast differentiation.
a–f, Csn1bGSKD (b), Csn2GSKD (c), Csn3GSKD (d), Csn7GSKD (e) and Nedd8GSKD (f) germaria contained four or more cystoblasts (arrowheads) in contrast to the one cystoblast in the Csn1aGSKD germarium (a). The ovals highlight GSCs. g, Quantification results from the independent knock-down experiment (from Fig. 4c) show that germline-specific knock down of Csn1b, Csn2, Csn3, Csn5, Csn6, Csn7 and Nedd8 significantly increased the number of germaria carrying three or more cystoblasts in comparison with the GFP knock-down negative control. Nedd8 knock down yielded the most severe differentiation defect. h–i, The bamΔ86/Csn5n (h′), bamΔ86/Csn3FS (h′′) and bamΔ86/csn7MB (h′′′) transheterozygous germaria had significantly more cystoblasts (arrowhead) plus two-cell pairs in their anterior region (bracket) than did the Csn5n (h) heterozygous germarium. Quantification results are shown in i. j, GSC quantification results show that Csn and bam transheterozygous ovaries did not contain significantly more GSCs than heterozygous Csn mutant ovaries. k–l′, Csn4k/+ (k) and Csn4n/+ (l) heterozygous germaria, as well as csn4k/+; bamΔ86/+ (k′) and csn4n/+; bamΔ86/+ (l′) transheterozygous germaria contained one cystoblast (arrowhead) or one two-cell pair (arrow). Ovals highlight GSCs. m, m′, Quantification results show that mutations in Csn4, Csn4EY, Csn4k and Csn4n suppressed the differentiation defect of the bam heterozygous mutant, despite Csn4 and bam transheterozygotes having more GSCs than bam heterozygotes.
Extended Data Figure 9 Bam interferes with the interaction between Csn4 and other Csn proteins.
a, Bam did not associate with Csn5, Csn6 and Csn7 when co-expressed in S2 cells in the presence of Csn4. The asterisk indicates a non-specific band. b, The presence of the Bam protein decreased the ability of Csn4 to co-IP with Csn5, Csn6 and Csn7 in S2 cells. c, The presence of the Bam protein did not affect the ability of Csn6 to co-IP with Csn5 and Csn7. d, Quantification results for three independent experiments shown in c.
Rights and permissions
About this article
Cite this article
Pan, L., Wang, S., Lu, T. et al. Protein competition switches the function of COP9 from self-renewal to differentiation. Nature 514, 233–236 (2014). https://doi.org/10.1038/nature13562
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13562
This article is cited by
-
Analysis of Drosophila melanogaster testis transcriptome
BMC Genomics (2018)
-
Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo
Nature Communications (2018)
-
Transcriptomic Analysis of Male Black Tiger Shrimp (Penaeus monodon) After Polychaete Feeding to Enhance Testicular Maturation
Marine Biotechnology (2017)
-
Inhibitory role of reactive oxygen species in the differentiation of multipotent vascular stem cells into vascular smooth muscle cells in rats: a novel aspect of traditional culture of rat aortic smooth muscle cells
Cell and Tissue Research (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.