Abstract
Receptor interacting protein kinase 1 (RIPK1) has an essential role in the signalling triggered by death receptors and pattern recognition receptors1,2. RIPK1 is believed to function as a node driving NF-κB-mediated cell survival and inflammation as well as caspase-8 (CASP8)-dependent apoptotic or RIPK3/MLKL-dependent necroptotic cell death. The physiological relevance of this dual function has remained elusive because of the perinatal death of RIPK1 full knockout mice3. To circumvent this problem, we generated RIPK1 conditional knockout mice, and show that mice lacking RIPK1 in intestinal epithelial cells (IECs) spontaneously develop severe intestinal inflammation associated with IEC apoptosis leading to early death. This early lethality was rescued by antibiotic treatment, MYD88 deficiency or tumour-necrosis factor (TNF) receptor 1 deficiency, demonstrating the importance of commensal bacteria and TNF in the IEC Ripk1 knockout phenotype. CASP8 deficiency, but not RIPK3 deficiency, rescued the inflammatory phenotype completely, indicating the indispensable role of RIPK1 in suppressing CASP8-dependent apoptosis but not RIPK3-dependent necroptosis in the intestine. RIPK1 kinase-dead knock-in mice did not exhibit any sign of inflammation, suggesting that RIPK1-mediated protection resides in its kinase-independent platform function. Depletion of RIPK1 in intestinal organoid cultures sensitized them to TNF-induced apoptosis, confirming the in vivo observations. Unexpectedly, TNF-mediated NF-κB activation remained intact in these organoids. Our results demonstrate that RIPK1 is essential for survival of IECs, ensuring epithelial homeostasis by protecting the epithelium from CASP8-mediated IEC apoptosis independently of its kinase activity and NF-κB activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Declercq, W., Vanden Berghe, T. & Vandenabeele, P. RIP kinases at the crossroads of cell death and survival. Cell 138, 229–232 (2009)
Ofengeim, D. & Yuan, J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nature Rev. Mol. Cell Biol. 14, 727–736 (2013)
Kelliher, M. A. et al. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity 8, 297–303 (1998)
Berger, S. B. et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480 (2014)
Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014)
Cho, Y. S. et al. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009)
He, S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 (2009)
Zhang, D. W. et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009)
Murphy, J. M. et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013)
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012)
Tenev, T. et al. The ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 43, 432–448 (2011)
Feoktistova, M. et al. cIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell 43, 449–463 (2011)
Dondelinger, Y. et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 20, 1381–1392 (2013)
Bertrand, M. J. et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 (2008)
Ea, C. K., Deng, L., Xia, Z. P., Pineda, G. & Chen, Z. J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 (2006)
O'Donnell, M. A., Legarda-Addison, D., Skountzos, P., Yeh, W. C. & Ting, A. T. Ubiquitination of RIP1 regulates an NF-κB-independent cell-death switch in TNF signaling. Curr. Biol. 17, 418–424 (2007)
Kajino-Sakamoto, R. et al. TGF-β-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J. Immunol. 185, 4729–4737 (2010)
Nenci, A. et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446, 557–561 (2007)
Günther, C. et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 (2011)
Welz, P. S. et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477, 330–334 (2011)
Vince, J. E. et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 36, 215–227 (2012)
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009)
Gentle, I. E. et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-κB and activation of caspase-8. J. Biol. Chem. 286, 13282–13291 (2011)
Vanlangenakker, N. et al. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 18, 656–665 (2011)
Oberst, A. et al. Catalytic activity of the caspase-8–FLIPL complex inhibits RIPK3-dependent necrosis. Nature 471, 363–367 (2011)
Kaiser, W. J. et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471, 368–372 (2011)
Moujalled, D. M. et al. In mouse embryonic fibroblasts, neither caspase-8 nor cellular FLICE-inhibitory protein (FLIP) is necessary for TNF to activate NF-κB, but caspase-8 is required for TNF to cause cell death, and induction of FLIP by NF-κB is required to prevent it. Cell Death Differ. 19, 808–815 (2012)
Dillon, C. P. et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202 (2014)
Rickard, J. A. et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 157, 1175–1188 (2014)
Kaiser, W. J. et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc. Natl Acad. Sci. USA 111, 7753–7758 (2014)
Dannoppel, M. et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature http://dx.doi.org/10.1038/nature13608 (this issue)
Rothe, J. et al. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature 364, 798–802 (1993)
Adachi, O. et al. Targeted disruption of the MYD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9, 143–150 (1998)
Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283 (2002)
El Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004)
Newton, K., Sun, X. & Dixit, V. M. Kinase RIP3 is dispensable for normal NF-κBs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and toll-like receptors 2 and 4. Mol. Cell. Biol. 24, 1464–1469 (2004)
Schoonjans, L. et al. Improved generation of germline-competent embryonic stem cell lines from inbred mouse strains. Stem Cells 21, 90–97 (2003)
Rodríguez, C. I. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature Genet. 25, 139–140 (2000)
Metzger, D. & Chambon, P. Site- and time-specific gene targeting in the mouse. Methods 24, 71–80 (2001)
Betz, U. A., Vosshenrich, C. A., Rajewsky, K. & Muller, W. Bypass of lethality with mosaic mice generated by Cre–loxP-mediated recombination. Curr. Biol. 6, 1307–1316 (1996)
Becker, C., Fantini, M. C. & Neurath, M. F. High resolution colonoscopy in live mice. Nature Protocols 1, 2900–2904 (2007)
Sato, T. & Clevers, H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2013)
Acknowledgements
We thank the Transgenic Mice Core Facility (IRC, VIB-UGent, Ghent) for their assistance. Villin-Cre, villin-Cre-ERT2 and RIPK3 knockout mice were provided by D. Gumucio (University of Michigan), S. Robine (Paris, France) and K. Newton and V. Dixit (Genentech), respectively. We thank A. Bredan for editing the manuscript, and P. De Bleser and M. Vuylsteke for statistical analysis. P.V. is senior full professor at Ghent University and holder of a Methusalem grant. L.V. is holder of a fellowship from Research Foundation Flanders (FWO). W.D. and G.V.L. have research professor positions at Ghent University. M.B. has a tenure track position in the Multidisciplinary Research Program of Ghent University (GROUP-ID). N.T. and V.G. are paid by the Methusalem grant, S.L. by a VIB grant and S.K. by a grant from the FWO. G.V.L. was supported by an FWO Odysseus Grant and by research grants from FWO, Foundation against Cancer and the Queen Elisabeth Medical foundation. Research in P.V.’s group is supported by Belgian grants (Interuniversity Attraction Poles, IAP 7/32), Flemish grants (Research Foundation Flanders, FWO G.0875.11, FWO G.0973.11, FWO G.0A45.12N, FWO G.0787.13N, G.0544.11N, G0C3114N and Methusalem grant BOF09/01M00709), Ghent University grants (MRP, GROUP-ID consortium), grant from the Foundation against Cancer (F94 and 2010-162) and grants from VIB. C.B. and C.G. received funding from the IZKF of the FAU Erlangen-Nürnberg and the DFG within the projects SPP1656, BE3686/2 and SFB796.
Author information
Authors and Affiliations
Contributions
N.T., L.V., M.J.M.B., W.D., G.v.L. and P.V. designed the study. N.T., L.V., M.J.M.B., L.D., T.D., M.S., S.K., V.G., S.L., C.G. and B.G. performed the experiments. N.T., L.V., M.J.M.B., A.G., W.D., G.v.L. and P.V. analysed the data. N.T., M.J.M.B., W.D., G.v.L. and P.V. wrote the manuscript. S.B.B., C.B., J.B. and P.J.G. provided reagents and scientific insight.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Generation of conditional Ripk1−/− mice using Cre–loxP technology.
a, Schematic representation of the targeting vector along with wild-type and mutant loci. Double homologous recombination between the wild-type Ripk1 allele and the targeting vector leads to entry of the neomycin positive selection marker and of extra ScaI (*) and PvuII (**) restriction sites in the Ripk1 allele and exclusion of the thymidine kinase (TK) negative selection marker, generating the Ripk1Nfl allele. Southern blot analysis reveals a 4.0-kb DNA fragment instead of a 10.3-kb fragment with the 5′ probe and a 5.3-kb instead of a 10.0-kb fragment with the 3′ probe for the Ripk1Nfl allele compared to the wild-type Ripk1. The Frt-flanked neomycin cassette was removed by crossing Ripk1Nfl mice with a Flp-deleter strain to generate a Ripk1 floxed allele (Ripk1fl). To ablate RIPK1, Ripk1fl/fl mice were crossed with Cre-deleter transgenic mice to induce germline recombination of the loxP sequences or with cell-type-specific Cre transgenic lines to induce tissue-specific recombination. The loxP interstitial exon 3 is looped out, disabling Ripk1 transcription. b, Southern blot analysis of five positive embryonic stem cell clones using a 5′ probe and a 3′ probe. MW, molecular weight; WT, wild type; Rec, recombinant. c. Ripk1+/+ and Ripk1Nfl/+ genotypes were distinguished by PCR using the primers indicated by arrows in a. d, MEF cells were prepared from Ripk1+/+, Ripk1+/− and Ripk1−/− embryos on embryonic day 14.5, and RIPK1 protein levels were analysed in lysates by western blot. β-Actin protein levels were used as a loading control.
Extended Data Figure 2 RIPK1 deficiency in MEFs results in defective MAPK and NF-κB activation associated with apoptosis induction upon TNF stimulation.
Ripk1−/− MEFs and control Ripk1+/+ MEFs were stimulated with 10 ng ml−1 of mTNF. a, Western blot analysis of NF-κB and MAPK activation in lysates sampled at different time points. b, RNA was analysed by qPCR for TNF, A20, IκBα and MCP1 expression (n = 3). Data are represented by mean ± s.e.m.
Extended Data Figure 3 Characterization of Ripk1IEC-KO mice.
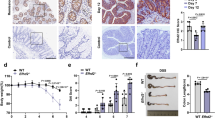
a, Birth and survival rates of all genotypes of offspring from selected breeding couples (Ripk1fl/fl × Ripk1fl/+ villin-creTg). *P < 0.05 by Fischer’s exact test. b, Genomic PCR analysis of Ripk1 alleles on DNA extracted from various tissues of Ripk1IEC-KO mice and control Ripk1fl/fl mice. c, Western blot detection of RIPK1 protein in small intestine and colon tissue of Ripk1IEC-KO and control Ripk1fl/fl mice. KO, knockout; bp, base pairs. d, PAS/AB staining of colons of Ripk1IEC-KO and Ripk1fl/fl mice (the same sample set as in Fig. 1c). e, Immunohistochemistry of activated CASP3 in the same sample set as in d.
Extended Data Figure 4 Characterization of older Ripk1IEC-KO mice.
a, Macroscopic images showing 22-week-old Ripk1fl/fl and Ripk1IEC-KO mice, and corresponding splenomegaly and colon shortening in Ripk1IEC-KO mice. b, Haematoxylin and eosin staining of Ripk1fl/fl (n = 3) and Ripk1IEC-KO (n = 3) mice at the ages of 6 weeks and 22 weeks. A representative image is shown.
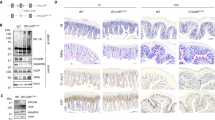
Extended Data Figure 5 Rescue of the inflammatory phenotype of Ripk1IEC-KO mice by antibiotic treatment.
a, Birth and survival rates (at weaning age) of all genotypes either treated or untreated with broad-spectrum antibiotics (ABX). The interaction between the genotype and the antibiotic treatment was assessed by analysis of deviance. b, Histopathological analysis by haematoxylin and eosin and immunohistochemical staining of Lysozyme P (LysP, Paneth cells) in small intestine samples from mice with or without antibiotic treatment: Ripk1fl/fl mice (n = 5), Ripk1IEC-KO mice without antibiotics (n = 5) and Ripk1IEC-KO mice with antibiotics (n = 7). c, TNF level detected by qPCR of RNA isolated from small intestine of antiobiotic-treated and non-treated Ripk1fl/fl and Ripk1IEC-KO mice (all n = 5). ***P = 0.0001 by two-tailed, two-way ANOVA. Data represent mean ± s.e.m. WT, wild type; KO, knockout. d, Antibiotic-treated Ripk1fl/fl (n = 3) and Ripk1iIEC-KO (n = 7), and non-treated Ripk1fl/fl (n = 3) and Ripk1iIEC-KO (n = 5) mice were subjected to RIPK1 deletion by daily treatment with 1 mg 4-OHT, and survival rate was monitored. ***P = 0.0007.
Extended Data Figure 6 Rescue of the lethal phenotype of Ripk1IEC-KO mice by MYD88 deficiency or TNFR1 deficiency.
a, Birth and survival rates (at weaning age) of offspring of all genotypes from selected breeding couples (Ripk1fl/fl Myd88−/− × Ripk1fl/+ villin-creTg Myd88−/−); Pearson’s χ2 test. b, Birth and survival rates (at weaning age) of offspring of all genotypes from selected breeding couples (Ripk1fl/fl Tnfr1−/− × Ripk1fl/+ villin-creTg Tnfr1−/−); Pearson’s χ2 test. c, Haematoxylin and eosin staining and TUNEL assay of Ripk1fl/fl Myd88−/− mouse and a littermate Ripk1IEC-KO Myd88−/− mouse small intestine. d, Haematoxylin and eosin staining and TUNEL assay of Ripk1fl/flTnfr1−/− mouse and a littermate Ripk1IEC-KOTnfr1−/− mouse small intestine. e, TNF levels of small intestine lysate from Ripk1fl/fl (n = 5), Ripk1IEC-KO(n = 5), Ripk1IEC-KO Myd88−/− (n = 4) and Ripk1IEC-KO Tnfr1−/− mouse (n = 6) determined by qPCR analysis.
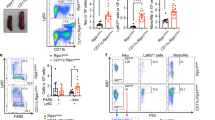
Extended Data Figure 7 Ripk1IEC-KO phenotype involves CASP8 but not RIPK3.
a, Survival rate of offspring of all genotypes from selected breeding couples (Ripk1fl/fl Casp8fl/fl × Ripk1fl/+ Casp8fl/+ villin-creTg) at weaning. b, An image of a 26-week-old Ripk1IEC-KO Casp8IEC-KO mouse and a littermate Ripk1fl/fl Casp8fl/fl mouse showing no detectable difference. All Ripk1IEC-KO Casp8IEC-KO mice at different ages (n = 6) were indistinguishable from littermate Ripk1fl/fl Casp8fl/fl mice (n = 8). c, Survival rate of offspring all of genotypes from selected breeding couples (Ripk1fl/flRipk3−/− × Ripk1fl/+ Ripk3−/− villin-creTg) at weaning showing significantly fewer Ripk1fl/fl Ripk3−/− mice surviving at weaning. d, An image of a 9-week-old Ripk1IEC-KO Ripk3−/− mouse and a littermate Ripk1fl/fl Ripk3−/− mouse. All Ripk1IEC-KORipk3−/− mice at different ages (n = 5) were smaller than littermate Ripk1fl/fl Casp8fl/fl mice (n = 14). e, Haematoxylin and eosin staining and TUNEL assay of small intestine of Ripk1IEC-KO, Ripk1fl/+ Casp8IEC-KO (indicated as Casp8IEC-KO), Ripk1IEC-KO Casp8IEC-KO, Ripk1IEC-KO Casp8IEC-KO and Ripk1IEC-KO Ripk3−/− mice. f, The number of necrotic cells per crypt was counted based on haematoxylin and eosin staining.
Extended Data Figure 8 RIPK1 kinase activity is not required for intestinal homeostasis.
Haematoxylin and eosin staining and TUNEL assay and immunohistochemical staining of active CASP3 (C3*) in small intestine samples from Ripk1fl/fl mice (n = 5), Ripk1IEC-KO mice (n = 5) and RIPK1-kinase-dead knock-in mice (Ripk1KD-KI, n = 5) at the age of 6−14 weeks. The images of 12-week-old mice are shown. Scale bars, 50 µm; black arrows indicate active CASP3 positivity. A magnified view is inserted in the left corner of the immunohistochemistry images.
Extended Data Figure 9 TNF-independent death of organoids and no direct cell death induction by TLR4 or TLR2 ligand.
Intestinal organoids were derived from small intestine crypt cells isolated from wild-type (WT) and Ripk1iIEC-KO mice. RIPK1 deletion (iKO) was induced in vitro by 4-OHT treatment (200 nM) for 20−24 h. a, Organoids were observed for up to 4 days after removal of 4-OHT. b, Organoids were treated with 10 ng ml−1 mTNF, Gram-negative bacterial lipopolysaccharide (TLR4 ligand, 100 µg ml−1), or Gram-positive bacterial lipoteichoic acid (TLR2 ligand, 100 µg ml−1) after removal of 4-OHT. Time-lapse confocal microscopic imaging of propidium iodide (PI)-stained organoids (n = 2). Scale bar, 100 µm. c, Image analysis of the percentage of propidium-iodide-positive nuclei in an experiment resembling those in b.
Extended Data Figure 10 Degradation of cFLIP in TNF-treated Ripk1−/− MEFs and Ripk1iIEC-KO organoids.
a, Protein lysates were prepared at designated times from Ripk1−/− MEFs treated with mTNF (10 ng ml−1) and analysed by western blot for NF-κB-inducing kinase (NIK), TRAF2, cIAP1 and cFLIP. b, Protein lysates were prepared at designated times from Ripk1iIEC-KO organoids treated with 4-OHT (200 nM) for 20 h before stimulation with mTNF (10 ng ml−1), and analysed by western blot for cIAP1. c, Protein lysates isolated from Ripk1KD-KI organoids were stimulated with mTNF (10 ng ml−1) in an independent experiment and analysed for RIPK1 and cFLIP. WT, wild type.
Supplementary information
Time-lapse confocal microscopic imaging of Ripk1iEC-KO organoids treated with vehicle and 10 ng/ml mTNF
Intestinal organoids were derived from small intestine crypt cells isolated from Ripk1iECK mice and treated with vehicle for 20–24 h, and subsequently with 10 ng/ml recombinant murine TNF (mTNF) 2 h after removal of vehicle. Cell death was visualized by incorporation of propidium iodide (PI)(3µM). Representative video frame. (MP4 2192 kb)
Time-lapse confocal microscopic imaging of Ripk1iEC-KO organoids treated with 4-OHT and 10 ng/ml mTNF
Intestinal organoids were derived from small intestinal crypt cells isolated from Ripk1iECK mice and RIPK1 was then deleted in vitro by 4-OHT treatment (200 nM) for 20–24 h. Organoids were subsequently treated with 10 ng/ml recombinant murine TNF (mTNF) 2 h after removal of 4-OHT. Cell death was visualized by incorporation of PI (3μM). Representative video frame. Supplementary videos 1 and 2 originate from the same experiment, and were repeated more than 3 times. (MP4 6126 kb)
Time-lapse confocal microscopic imaging of WT organoids treated with 10 ng/ml mTNF
Intestinal organoids were derived from small intestinal crypt cells isolated from WT mice, treated with 10 ng/ml recombinant murine TNF (mTNF). Cell death was visualized by incorporation of PI (3μM). Representative video frame. (MP4 10633 kb)
Time-lapse confocal microscopic imaging of Ripk1iEC-KO organoids treated with 4-OHT and 10 ng/ml mTNF.
Intestinal organoids were derived from small intestinal crypt cells isolated from Ripk1iECKO mice, and RIPK1 was deleted in vitro by 4-OHT treatment (200 nM) for 20–24 h. Organoids were subsequently treated with 10 ng/ml recombinant murine TNF (mTNF) 2 h after removal of 4-OHT. Cell death was visualized by incorporation of PI (3μM). Representative video frame. (MP4 14749 kb)
Time-lapse confocal microscopic imaging of Ripk1KD-KI organoids treated with 10 ng/ml mTNF
Intestinal organoids were derived from small intestinal crypt cells isolated from Ripk1KD-KI mice, and treated with 10 ng/ml recombinant murine TNF (mTNF). Cell death was visualized by incorporation of PI (3μM). Representative movie frame. Supplementary videos 3-5 were recorded in a single experiment, and repeated 2 times. (MP4 10171 kb)
Time-lapse confocal microscopic imaging of WT organoids treated with 4-OHT
Intestinal organoids were derived from small intestinal crypt cells isolated from WT mice are treated with 4-OHT (200 nM) for 20–24 h. Imaging started 2h after the removal of 4OHT. Cell death was visualized by incorporation (MP4 11909 kb)
Time-lapse confocal microscopic imaging of Ripk1iEC-KO organoids treated with 4-OHT
Intestinal organoids were derived from small intestinal crypt cells isolated from Ripk1iEOK mice are treated with 4-OHT (200 nM) for 20–24 h. Imaging started 2h after the removal of 4OHT. Cell death was visualized by incorporation of PI (3μM). Representative movie frame. Supplementary videos 3, 4, 5, 6 and 7 were recorded in a single experiment, and repeated 2 times. (MP4 9296 kb)
Rights and permissions
About this article
Cite this article
Takahashi, N., Vereecke, L., Bertrand, M. et al. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature 513, 95–99 (2014). https://doi.org/10.1038/nature13706
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13706
This article is cited by
-
Mediators of necroptosis: from cell death to metabolic regulation
EMBO Molecular Medicine (2024)
-
Immunogenic cell death in cancer: targeting necroptosis to induce antitumour immunity
Nature Reviews Cancer (2024)
-
PANoptosis: a potential new target for programmed cell death in breast cancer treatment and prognosis
Apoptosis (2024)
-
Regulated cell death pathways and their roles in homeostasis, infection, inflammation, and tumorigenesis
Experimental & Molecular Medicine (2023)
-
RIPK1 plays a crucial role in maintaining regulatory T-Cell homeostasis by inhibiting both RIPK3- and FADD-mediated cell death
Cellular & Molecular Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.