Abstract
The blood system is sustained by a pool of haematopoietic stem cells (HSCs) that are long-lived due to their capacity for self-renewal. A consequence of longevity is exposure to stress stimuli including reactive oxygen species (ROS), nutrient fluctuation and DNA damage1,2. Damage that occurs within stressed HSCs must be tightly controlled to prevent either loss of function or the clonal persistence of oncogenic mutations that increase the risk of leukaemogenesis3,4. Despite the importance of maintaining cell integrity throughout life, how the HSC pool achieves this and how individual HSCs respond to stress remain poorly understood. Many sources of stress cause misfolded protein accumulation in the endoplasmic reticulum (ER), and subsequent activation of the unfolded protein response (UPR) enables the cell to either resolve stress or initiate apoptosis5,6. Here we show that human HSCs are predisposed to apoptosis through strong activation of the PERK branch of the UPR after ER stress, whereas closely related progenitors exhibit an adaptive response leading to their survival. Enhanced ER protein folding by overexpression of the co-chaperone ERDJ4 (also called DNAJB9) increases HSC repopulation capacity in xenograft assays, linking the UPR to HSC function. Because the UPR is a focal point where different sources of stress converge, our study provides a framework for understanding how stress signalling is coordinated within tissue hierarchies and integrated with stemness. Broadly, these findings reveal that the HSC pool maintains clonal integrity by clearance of individual HSCs after stress to prevent propagation of damaged stem cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
11 June 2014
Superscript plus and minus symbols were incorrect in the labels for Fig. 3d and have been fixed.
References
Geiger, H., de Haan, G. & Florian, M. C. The ageing haematopoietic stem cell compartment. Nature Rev. Immunol. 13, 376–389 (2013)
Yahata, T. et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 118, 2941–2950 (2011)
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014)
Rossi, D. J., Jamieson, C. H. M. & Weissman, I. L. Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008)
Rutkowski, D. T. & Kaufman, R. J. A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28 (2004)
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011)
Majeti, R., Park, C. Y. & Weissman, I. L. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1, 635–645 (2007)
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature Immunol. 11, 585–593 (2010)
Notta, F. et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221 (2011)
Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nature Immunol. 14, 756–763 (2013)
DuRose, J. B., Tam, A. B. & Niwa, M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol. Biol. Cell 17, 3095–3107 (2006)
Tabas, I. & Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nature Cell Biol. 13, 184–190 (2011)
Lu, P. D., Harding, H. P. & Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 (2004)
Vattem, K. M. & Wek, R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA 101, 11269–11274 (2004)
Palam, L. R., Baird, T. D. & Wek, R. C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 (2011)
Marciniak, S. J. et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 (2004)
Han, J. et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biol. 15, 481–490 (2013)
Novoa, I., Zeng, H., Harding, H. P. & Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153, 1011–1022 (2001)
Boyce, M. et al. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307, 935–939 (2005)
Axten, J. M. et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 55, 7193–7207 (2012)
Shen, Y., Meunier, L. & Hendershot, L. M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947–15956 (2002)
Lai, C. W., Otero, J. H., Hendershot, L. M. & Snapp, E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J. Biol. Chem. 287, 7969–7978 (2012)
Kurisu, J. et al. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells 8, 189–202 (2003)
Rouault-Pierre, K. et al. HIF-2α protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell 13, 549–563 (2013)
Milyavsky, M. et al. A distinctive DNA damage response in human hematopoietic Stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell 7, 186–197 (2010)
Heijmans, J. et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013)
Wey, S., Luo, B. & Lee, A. S. Acute inducible ablation of GRP78 reveals its role in hematopoietic stem cell survival, lymphogenesis and regulation of stress signaling. PLoS ONE 7, e39047 (2012)
Warner, J. K. et al. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia 19, 1794–1805 (2005)
Amendola, M., Venneri, M. A., Biffi, A., Vigna, E. & Naldini, L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nature Biotechnol. 23, 108–116 (2005)
The MGC Project Team The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res. 14, 2121–2127 (2004)
Gentner, B. et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nature Methods 6, 63–66 (2009)
Subach, O. M. et al. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15, 1116–1124 (2008)
van Galen, P. et al. Reduced lymphoid lineage priming promotes human hematopoietic stem cell expansion. Cell Stem Cell 14, 94–106 (2014)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005)
Lopes, C. T. et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics 26, 2347–2348 (2010)
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001)
Lin, J. H. et al. IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944–949 (2007)
Harding, H. P., Zhang, Y. & Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 (1999)
Acknowledgements
We thank D. Ron and D. Rubinsztein for critical assessment of this work; all members of the Dick laboratory, especially O. I. Gan and E. Lechman, for experimental support and advice; A. Khandani, P. Penttilä and the SickKids-UHN flow facility for technical support; N. Jamal for providing bone marrow samples; J. Heijmans for providing the pLV-ca-GADD34 vector; and L. Naldini for providing the MA1 vector. Work in the Dick laboratory is supported by grants from the Canadian Institutes for Health Research, Canadian Cancer Society, Terry Fox Foundation, Genome Canada through the Ontario Genomics Institute, Ontario Institute for Cancer Research with funds from the province of Ontario, a Canada Research Chair and the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. A.R.G. and D.G.K. are supported by Leukemia and Lymphoma Research, Cancer Research UK, the Kay Kendall Leukaemia Fund, the NIHR Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia & Lymphoma Society of America. J.C.G. and T.F. are supported by Arthritis Research UK. S.J.M. and J.E.C. are supported by the Medical Research Council (UK). B.G.W. is supported by the Terry Fox New Frontiers Research Program (PPG09-020005), the Ontario Institute for Cancer Research, and the Canadian Institute for Health Research (CIHR grant 201592).
Author information
Authors and Affiliations
Contributions
P.v.G., A.K. and J.E.D. designed the study and analysed and interpreted the data. P.v.G., A.K., N.M., D.G.K., T.F., J.E.C., S.X., K.H. and E.W. performed experiments. E.L. and K.E. performed bioinformatic analyses. S.J.M., J.C.G., A.R.G. and B.G.W. supervised specific experiments. P.v.G. wrote the paper. A.K. and J.E.D. revised the paper. J.E.D. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
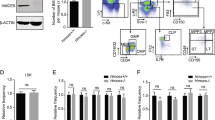
Extended Data Figure 1 Expression analysis of UPR-related genes.
a, Enrichment of UPR-related genes in human HSCs compared to progenitors. CD49f+ HSC-enriched genes were analysed for GO category overrepresentation. Node size represents the number of genes; white, yellow and orange colour correspond to FDR <0.15, <0.1 and <0.01. b, Simplified scheme illustrating UPR signalling events. Three branches of the UPR are activated upon ER stress: IRE1, PERK and ATF6. IRE1 splices cytosolic XBP1 mRNA to enable translation of the XBP1s transcription factor, which upregulates chaperones and ER-associated degradation (ERAD) machinery to resolve ER stress37,38. PERK initiates a different branch of the UPR through phosphorylation of eIF2α, which attenuates global protein synthesis, thus permitting time to restore ER homeostasis39. Prolonged ER stress leads to PERK signalling-mediated upregulation of the proapoptotic transcription factor CHOP and its target GADD34. GADD34 dephosphorylates eIF2α leading to restoration of global protein translation. However, if ER stress is not resolved, GADD34 upregulation can lead to further accumulation of misfolded proteins, oxidative stress and apoptosis16. Yellow highlighted arrows indicate transcriptional regulation. c, Amplification curves of qPCR reactions for UPR-related genes. Fluorescence signal during 40 cycles of qPCR reactions on cord-blood-derived cDNA is shown for a representative experiment. Green line indicates threshold that was used to calculate mRNA quantity. d, Dissociation curves were generated to check for the presence of aspecific amplicons or primer dimers, which would be visible as additional peaks. Each line represents the dissociation curve of one qPCR reaction, colours indicate different genes. e, Slopes and R2 values of standard curves are shown for a representative experiment. These values were calculated separately for each experiment, based on a cDNA dilution series. c–e were performed using SDS v2.3 software. f, Agarose gel analysis of qPCR amplicons. qPCR reactions were run on a 3% agarose gel to check for reaction specificity: nonspecific amplicons would be visible as additional bands. The expected product size is shown above the gel; the ladder sizes are indicated on the right. g, Adult bone marrow cells were sorted into HSPC and progenitor fractions. mRNA levels for CHOP and ERDJ4 were measured by qPCR. Results are shown as mean ± s.e.m. of n = 5 bone marrow samples. ****P < 0.0001.
Extended Data Figure 2 Differential response of HSPCs and progenitors to ER-stress-inducing agents.
a, b, HSPC and progenitor fractions were sorted and plated in the presence of (a) thapsigargin or (b) tunicamycin. mRNA was isolated after 0.5, 1, 6, 16 and 40 h and expression levels of GRP78, ERDJ4, GADD34 and ATF4 were assessed by qPCR. The DMSO-treated controls were the same between a and b. Data are shown as mean ± s.e.m. of n = 3 cord blood samples, P value was calculated based on fold change of treated over DMSO control cells and indicates differential response between HSPCs and progenitors. c, Adult bone marrow HSPCs and progenitors were sorted and plated in the presence of tunicamycin. After 16 h, mRNA was isolated and expression levels of CHOP, ERDJ4 and GRP94 were assessed by qPCR. Data are shown as mean ± s.e.m. of n = 5 bone marrow samples.
Extended Data Figure 3 Survival of HSCs is lower compared to progenitors after tunicamycin, but not thapsigargin treatment.
a, Thapsigargin has similar toxicity for sorted HSC and progenitor fractions. Sorted HSCs and progenitors were plated in TSGF6 culture conditions in the presence of thapsigargin or DMSO control. Symbols represent viable cell counts of individual samples where fractions are connected by a black line; the blue line indicates mean ± s.e.m. of n = 7 cord blood samples. b, c, Reduced clonogenic capacity of sorted HSCs compared to progenitors after tunicamycin treatment. Total colony number is shown in Fig. 2c. Here, data are separated into colony types based on morphological appearance. Data are shown as mean ± s.e.m. of n = 4 cord blood samples. G, granulocyte; M, macrophage; GM, granulocyte/macrophage; BFU, erythroid burst forming unit; mix, multilineage. d, HSCs have lower survival compared to progenitors after tunicamycin treatment, even after cell cycle induction. Sorted HSC and progenitor fractions were plated in TSGF6 culture conditions with double cytokine concentrations for 72–96 h to induce G0 exit of the HSC fraction33. Then, cells were plated in the presence of tunicamycin. Viable cell counts as a percentage of DMSO controls are shown. Symbols represent individual samples where fractions are connected by a black line; the blue line indicates mean ± s.e.m. of n = 5 cord blood samples at 0.6 μg ml−1 and n = 3 cord blood samples at 3 μg ml−1 tunicamycin. e, f, Increased apoptosis of HSCs compared to progenitors after tunicamycin treatment. e, Cord blood cells were plated with tunicamycin and stained for primitive surface markers, Annexin-V and Sytox. Representative flow plots are shown. f, Sorted HSCs and progenitors were plated in the presence of tunicamycin. The percentage of viable Annexin-V− cells after 40 h compared to DMSO controls is shown as mean ± s.e.m. of n = 4 cord blood samples. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Figure 4 ATF4 reporter enables visualization of increased ATF4 translation after tunicamycin treatment.
a, ATF4 reporter validation. Two upstream ORFs (uORFs) that are 5′ of the ATF4 coding sequence in the ATF4 mRNA ensure more efficient translation of ATF4 when eIF2α phosphorylation levels are high13,14. A bidirectional lentiviral vector was constructed that gives constitutive expression of TagBFP to mark transduced cells. In the other direction, the SFFV promoter drives expression of the 5′ end of the ATF4 mRNA which fuses with a GFP reporter gene 3′ of the termination codon of uORF2. HeLa cells were transduced with pSMALB-ATF4.5rep (referred to as ATF4 reporter) and treated with tunicamycin. After 30 h, GFP fluorescence was read out by flow cytometry. Histogram plots show n = 2 technical duplicates (two black lines for DMSO control, two red lines for tunicamycin treatment). b, c, Reporter fluorescence depends on uORFs. HeLa cells were transduced and treated with tunicamycin. As expected, ATF4–GFP translation is (b) repressed in the negative control that has a mutated uORF1 start codon and (c) constitutively high in the positive control with mutated start codons for both uORFs13. Histogram plots show n = 2 technical duplicates. d, ATF4 reporter-transduced cord blood cells were treated with tunicamycin and increasing doses of the PERK inhibitor GSK2606414. The transgene ratio (TGR) is shown as mean ± s.e.m. of n = 6 cord blood samples (except at 600 nM, n = 3 cord blood samples). *P < 0.05, **P < 0.01, ****P < 0.0001.
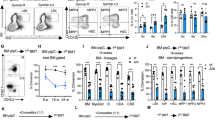
Extended Data Figure 5 Modulation of UPR-associated genes affects haematopoietic stem and progenitor cells in vivo.
a, Analysis of haematopoietic stem and progenitor cell frequencies in Chop–/– mice. Flow cytometry was performed on mouse bone marrow (Extended Data Table 2). Bars show the absolute cell production in each population from wild-type or Chop–/– mice. Data are shown as mean ± s.d. of n = 5 mice per group. b, Viability analysis of stem and progenitor cell populations in Chop–/– mice. The percentage of viable Annexin-V–7-AAD– cells within the HSC-enriched LSK and Lin–Sca-1–c-Kit+ progenitor fractions was assessed by flow cytometry. Data are shown as mean ± s.d. of technical duplicates of n = 5 mice per group. c, ERDJ4OE cells show increased survival after tunicamycin treatment. The haematopoietic TEX cell line28 was transduced with SFFV-Ctrl or SFFV-ERDJ4OE lentiviral vectors and plated in the presence of 0.6 μg ml−1 tunicamycin (SFFV refers to lentiviral promoter driving transgene expression). After 48 h, the number of transduced cells compared to DMSO-treated controls was determined by automated counting of GFP+ cells. Data are shown as mean ± s.d. of n = 3 independent experiments, P value was calculated using a paired t-test. d, Tunicamycin-induced apoptosis is reduced by ERDJ4OE. Cells from c were analysed for Annexin-V and cleaved caspase-3 expression by flow cytometry. Data are shown as mean ± s.d. of n = 3 independent experiments, P values were calculated using paired t-tests. e, ERDJ4OE endows cord blood cells with a competitive advantage over untransduced cells. Three cord blood pools (Exp. 1–3) were transduced with PGK-Ctrl or PGK-ERDJ4OE lentiviruses and injected into 5 mice each. Dashed line indicates GFP% after transduction (day 0); solid line indicates median GFP% of the human CD45+ graft in the injected femur of xenografted mice (20 weeks). Every symbol represents one mouse. f, Similar expansion of PGK-Ctrl and PGK-ERDJ4OE transduced cord blood cells in vitro. Three cord blood pools (Exp. 1–3) were transduced with PGK-Ctrl or PGK-ERDJ4OE lentivirus and expanded for 10 days in liquid culture. Total population doublings of transduced GFP+ cells is shown. g, ERDJ4OE increases HSC output. After liquid culture, GFP+ cells from f were sorted and injected at high and low cell doses, indicated below the x axis. Total human CD45+GFP+ engraftment in the injected femur after 10 weeks is shown. P values were calculated using the Mann–Whitney U-test. Every symbol represents one mouse, line shows median. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 6 Lineage differentiation, progenitor cell frequencies, homing, and serial transplantability are maintained following ERDJ4OE.
a, PGK-ERDJ4OE-transduced cord blood maintains multilineage differentiation potential in vivo. Left: gating scheme to assess differentiation of the human graft in mouse bone marrow. Representative flow plots show quantification of CD45+CD19+ B cells, CD45+CD33+ monocytes and granulocytes, and CD45−GlyA+ erythroid cells within the GFP+ graft. Right: the differentiation of transduced cord blood cells was assessed in the peripheral blood (PB) at 10 and 20 weeks and in the injected (RF) and non-injected (LF) femur at 20 weeks after transplantation. Results are shown as mean ± s.e.m. of n = 15 mice representing n = 3 cord blood samples. b, ERDJ4OE does not cause aberrant expansion of stem or progenitor cell fractions. To assess the distribution of human stem and progenitor cells, lineage+ and mouse cells were depleted from xenografted mouse bone marrow. The remaining human lineage– cells were analysed by flow cytometry. Left: gating scheme to assess differentiation into HSC, MPP, MLP, CMP/MEP and B/NK/GMP fractions (Extended Data Table 1). Right: the frequency of human stem and progenitor cells within the human CD45+GFP+ graft was assessed 20 weeks after transplantation of transduced cord blood cells. Results are shown as mean ± s.e.m. of n = 3 cord blood samples. c, Homing capacity to the non-injected bone marrow is not altered by ERDJ4OE. Transduced cord blood cells were expanded for 12 days in liquid culture conditions and 1–1.6 ×106 cells were transplanted per mouse. After 19 h, mice were euthanized to assess human CD45+GFP+ cell homing to the non-injected femur. Results were normalized to transduction efficiency. Every symbol represents one mouse, results of n = 3 cord blood samples are shown with 2 mice per group each; line shows median. d, Frequency of functional human HSCs in vivo is maintained with ERDJ4OE. Cord blood cells were transduced and injected into primary mice. After 10 weeks, mice were killed and transduced GFP+ cells were sorted from their bone marrow. Thirty thousand to one million cells were re-transplanted into secondary mice for serial LDA. After 10 weeks, the bone marrow of secondary mice was assessed for human CD45+GFP+ engraftment; mice were scored as positive if the engraftment level was >0.01%. Data from n = 3 cord blood samples was pooled.
Rights and permissions
About this article
Cite this article
van Galen, P., Kreso, A., Mbong, N. et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272 (2014). https://doi.org/10.1038/nature13228
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13228
This article is cited by
-
Tissue mosaicism following stem cell aging: blood as an exemplar
Nature Aging (2024)
-
MITOL deficiency triggers hematopoietic stem cell apoptosis via ER stress response
The EMBO Journal (2024)
-
EVA1A regulates hematopoietic stem cell regeneration via ER-mitochondria mediated apoptosis
Cell Death & Disease (2023)
-
Interlukin-4 weakens resistance to stress injury and megakaryocytic differentiation of hematopoietic stem cells by inhibiting Psmd13 expression
Scientific Reports (2023)
-
Hydrogel-based microenvironment engineering of haematopoietic stem cells
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.