Abstract
PINK1 (PTEN induced putative kinase 1) and PARKIN (also known as PARK2) have been identified as the causal genes responsible for hereditary recessive early-onset Parkinsonism1,2. PINK1 is a Ser/Thr kinase that specifically accumulates on depolarized mitochondria, whereas parkin is an E3 ubiquitin ligase that catalyses ubiquitin transfer to mitochondrial substrates3,4,5. PINK1 acts as an upstream factor for parkin6,7 and is essential both for the activation of latent E3 parkin activity8 and for recruiting parkin onto depolarized mitochondria8,9,10,11,12. Recently, mechanistic insights into mitochondrial quality control mediated by PINK1 and parkin have been revealed3,4,5, and PINK1-dependent phosphorylation of parkin has been reported13,14,15. However, the requirement of PINK1 for parkin activation was not bypassed by phosphomimetic parkin mutation15, and how PINK1 accelerates the E3 activity of parkin on damaged mitochondria is still obscure. Here we report that ubiquitin is the genuine substrate of PINK1. PINK1 phosphorylated ubiquitin at Ser 65 both in vitro and in cells, and a Ser 65 phosphopeptide derived from endogenous ubiquitin was only detected in cells in the presence of PINK1 and following a decrease in mitochondrial membrane potential. Unexpectedly, phosphomimetic ubiquitin bypassed PINK1-dependent activation of a phosphomimetic parkin mutant in cells. Furthermore, phosphomimetic ubiquitin accelerates discharge of the thioester conjugate formed by UBCH7 (also known as UBE2L3) and ubiquitin (UBCH7∼ubiquitin) in the presence of parkin in vitro, indicating that it acts allosterically. The phosphorylation-dependent interaction between ubiquitin and parkin suggests that phosphorylated ubiquitin unlocks autoinhibition of the catalytic cysteine. Our results show that PINK1-dependent phosphorylation of both parkin and ubiquitin is sufficient for full activation of parkin E3 activity. These findings demonstrate that phosphorylated ubiquitin is a parkin activator.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998)
Valente, E. M. et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004)
Narendra, D., Walker, J. E. & Youle, R. Mitochondrial quality control mediated by PINK1 and Parkin: links to Parkinsonism. Cold Spring Harb. Perspect. Biol. 4, a011338 (2012)
Corti, O., Lesage, S. & Brice, A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 91, 1161–1218 (2011)
Exner, N., Lutz, A. K., Haass, C. & Winklhofer, K. F. Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J. 31, 3038–3062 (2012)
Clark, I. E. et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006)
Park, J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006)
Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 (2010)
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010)
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 12, 119–131 (2010)
Vives-Bauza, C. et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA 107, 378–383 (2010)
Ziviani, E., Tao, R. N. & Whitworth, A. J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA 107, 5018–5023 (2010)
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012)
Shiba-Fukushima, K. et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2, 1002 (2012)
Iguchi, M. et al. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J. Biol. Chem. 288, 22019–22032 (2013)
Chaugule, V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011)
Zheng, X. & Hunter, T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 23, 886–897 (2013)
Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K. & Koike, T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteom. 5, 749–757 (2006)
Gautier, C. A., Kitada, T. & Shen, J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl Acad. Sci. USA 105, 11364–11369 (2008)
Okatsu, K. et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nature Commun. 3, 1016 (2012)
Okatsu, K. et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288, 36372–36384 (2013)
Trempe, J. F. et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 (2013)
Lazarou, M. et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172 (2013)
Ciehanover, A., Hod, Y. & Hershko, A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. 1978. Biochem. Biophys. Res. Commun. 81, 1100–1105 (1978)
Spence, J., Sadis, S., Haas, A. L. & Finley, D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 (1995)
Riley, B. E. et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nature Commun. 4, 1982 (2013)
Wauer, T. & Komander, D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 (2013)
Spratt, D. E. et al. A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nature Commun. 4, 1983 (2013)
Schapira, A. H. Complex I: inhibitors, inhibition and neurodegeneration. Exp. Neurol. 224, 331–335 (2010)
Tsuchiya, H., Tanaka, K. & Saeki, Y. The parallel reaction monitoring method contributes to a highly sensitive polyubiquitin chain quantification. Biochem. Biophys. Res. Commun. 436, 223–229 (2013)
Acknowledgements
This work was supported by JSPS KAKENHI grant numbers 23687018 (to N.M.), 23-6061 (to K.O.), 21000012 (to K.T.), 24657072 and 22227003 (to Y.T. and T.E.); MEXT KAKENHI grant numbers 24111557 and 25112522 (to N.M.), 24112008 (to Y.S.); the Tomizawa Jun-ichi and Keiko Fund (to N.M.); JST CREST (to T.E.); Platform for Drug Discovery, Informatics, and Structural Life Science from MEXT (to T.H.); startup funds from McGill University and the GRASP from the FRSQ (to J.-F.T.); an Operating Grant from the CIHR and a Chercheur National Award from the FRSQ (to E.A.F.); and the Takeda Science Foundation (to H.K., K.T. and N.M.). We thank N. Croteau and S. Rasool for technical assistance, T. Ueno for anti-FoF1 antibody, and D. Finley for yeast strain SUB328.
Author information
Authors and Affiliations
Contributions
F.K., K.O., H.K., E.G., J.-F.T. and M.K. carried out biochemical and immunoblot experiments. T.H. performed computational modelling. Y.T., Y.K., Y.S. and T.E. performed yeast experiments. F.K., E.G. and M.K. performed immunocytochemistry. F.K., H.T., H.Y., J.-F.T., E.A.F. and Y.S. performed mass spectrometric analysis. K.T. and N.M. designed the research and analysed the data. N.M. wrote the manuscript with help and supervision from K.T. All authors contributed to data analysis and preparation of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Ubiquitin Ser 65 is phosphorylated by depolarized mitochondria.
a, Sequence comparison between ubiquitin and the parkin UBL domain. Grey asterisks, identical amino acid residues. b, When the same samples as Fig. 1b were subjected to non-Phos-tag PAGE, no retarded-mobility form of ubiquitin was observed. Black asterisk, cross-reacting band. c, Recombinant SUMO-1 was incubated with depolarized mitochondria prepared from PINK1-expressing HeLa cells in vitro. Red asterisk, phosphorylated ubiquitin as a positive control under the same experimental conditions. d, Mass spectrometric (MS) analysis of the tryptic phosphopeptide from ubiquitin obtained in Fig. 1b. e, f, When the same samples as in Fig. 1c (e) and Fig. 1d (f) were subjected to non Phos-tag PAGE, no retarded-mobility form of ubiquitin was observed. g, The portion of the gel corresponding to the low-molecular-weight fraction of whole-cell extracts used for MS analysis of Fig. 1e. h, Whole-cell lysates of PINK1-expressing HeLa cells ± CCCP treatment were subjected to SDS-PAGE and the high- (>55 kDa), middle- (14–55 k) and low- (<14 k) molecular-weight fractions were collected for MS analysis of Extended Data Fig. 1i. i, Using AQUA peptides as standards, the absolute quantities of the ESTLHLVLR and EpSTLHLVLR peptides in the high, middle and total fractions were determined in three experiments. Error bars represent mean ± s.e.m. and statistical significance was calculated using a one-tailed paired t-test.
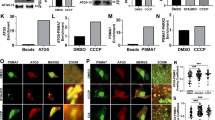
Extended Data Figure 2 PINK1 is essential for ubiquitin phosphorylation.
a, Results of the control experiments for Fig. 2a. The same samples as Fig. 2a were subjected to non-Phos-tag PAGE to confirm that no retarded-mobility form of ubiquitin was observed. b, PINK1 was immunoprecipitated and its purity was confirmed by immunoblot analysis with antibodies against other mitochondrial membrane proteins. c, Results of the control experiments for Fig. 2b. The same samples as Fig. 2b were subjected to non Phos-tag PAGE to confirm that no retarded-mobility form of ubiquitin was observed. d, Immunoblot analysis to confirm PINK1 knockdown. HeLa cells grown in light or heavy SILAC media were treated with non-target (NT) or PINK1 siRNA, respectively, and were subjected to immunoblot analysis with an anti-PINK1 antibody.
Extended Data Figure 3 Mass-spectrometry-based absolute quantification of phosphorylated ubiquitin in intact HeLa cells.
Whole-cell lysates of intact HeLa cells ± CCCP treatment were subjected to SDS-PAGE, and the high- (>55 k), middle- (14–55 k), or low- (<14 k) molecular-weight fractions were collected. The absolute quantities of the ESTLHLVLR and EpSTLHLVLR peptides in the high, middle and total fractions were then determined using AQUA peptides as standards in three experiments. Error bars represent mean ± s.e.m. and statistical significance was calculated using a one-tailed paired t-test. Because the signal intensity is low, the original data and chromatogram of EpSTLHLVLR peptide are also shown. The light signal (derived from endogenous EpSTLHLVLR peptide), which shows a similar ionized pattern and the identical elution time as the heavy signal (derived from internal AQUA EpSTLHLVLR control peptide), was observed only in CCCP-pretreated cells (red arrows).
Extended Data Figure 4 Subcellular localization of phosphomimetic or phosphorylation-deficient ubiquitin under various experimental conditions.
a, Phosphomimetic ubiquitin(Ser65Asp) stimulates the E3 activity of parkin in the absence of mitochondrial localization. Example figures indicative of cytoplasmic localization of both phosphomimetic parkin and ubiquitin are shown. Mitochondrial localization was 0% of 100 cells in two independent experiments. Bars indicate 10 µm unless otherwise specified. b, Mitochondrial accumulation of phosphomimetic or phosphorylation-deficient ubiquitin depending on parkin and CCCP treatment (3 h). c, Phosphomimetic ubiquitin(Ser65Asp) is conjugated to the parkin substrate mitofusin-2. Red bars indicate ubiquitination of mitofusin-2 with phosphomimetic Flag-ubiquitin (lane 2) or phosphorylation-deficient Flag-ubiquitin (lane 4).
Extended Data Figure 5 PINK1 was inactivated following heat treatment.
Autoubiquitination of GFP–parkin in cell extracts was reconstituted when incubated with depolarized mitochondria (lane 1), whereas heat pre-treatment of the mitochondria abolished autoubiquitination (lane 2). Because the parkin-activating potency of depolarized mitochondria was lost by heat treatment (at 90 °C for 10 min), mitochondrial proteins including PINK1 were likely inactivated by the aforementioned conditions.
Extended Data Figure 6 Reconstitution of parkin activation by PINK1 in yeast cells.
A plasmid encoding GFP–parkin or a pathogenic mutant GFP–parkin(Thr240Arg) under the control of a constitutive promoter and a plasmid encoding OM-PINK1 (40 amino acids at the N terminus of yeast mitochondrial outer-membrane protein Om45 were fused to the N terminus of PINK1 lacking the genuine 120 amino acids at the N terminus) under the control of a galactose (Gal)-inducible promoter were co-transfected into yeast cells. The yeast cell lysates were collected and subjected to immunoblot analysis with an anti-parkin or anti-PINK1 antibody. As is the case with mammalian cells, a putative autoubiquitination signal was observed with GFP–parkin in accordance with PINK1 mitochondrial accumulation (lanes 1–4), whereas the PINK1 kinase dead (KD) mutation (lanes 5–8) or pathogenic parkin Thr240Arg mutation (lanes 9–12) hindered it.
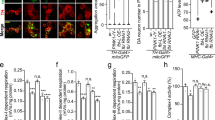
Extended Data Figure 7 In vitro charging and discharging assay of UBCH7.
a, Charging assay of UBCH7 using both phosphorylated and non-phosphorylated ubiquitin. Samples prepared as in Fig. 4f were resolved on SDS-PAGE using standard (non-Phos-tag) gel. b, Discharging assay of UBCH7 using wild-type, phosphomimetic and phosphorylated ubiquitin in the presence of parkin. His-UBCH7 was charged with N-terminal Fluorescein-labelled ubiquitin, and then purified His-UBCH7∼FluoUb (1 µM) was incubated with wild-type or mutant GST–parkin (0.5 µM) in the presence of 10 µM His-Ub wild-type, Ser65Asp or phospho-HisUb. Signal derived from Fluorescein-labelled ubiquitin was imaged using the fluorescent scanner. Fluorescence strength of His-UBCH7∼FluoUb bands was quantified in three experiments, and were subjected to statistical analysis as shown in Fig. 4g.
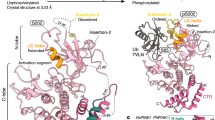
Extended Data Figure 8 Structural modelling of the fully activated parkin.
Based on the structure of the inactive, autoinhibited parkin (Model a), two possible models of fully activated parkin with higher accessibility to catalytic cysteine (Cys 431 in the RING2 domain) were generated. Binding of the phosphorylated UBL domain (Model b) or phosphorylated ubiquitin (Model c) to the RING0 domain supports a structural change in the RING2 domain. It is possible that Model b is an intermediate for Model c, or vice versa.
Rights and permissions
About this article
Cite this article
Koyano, F., Okatsu, K., Kosako, H. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). https://doi.org/10.1038/nature13392
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13392
This article is cited by
-
Optineurin provides a mitophagy contact site for TBK1 activation
The EMBO Journal (2024)
-
Activation of Ca2+ phosphatase Calcineurin regulates Parkin translocation to mitochondria and mitophagy in flies
Cell Death & Differentiation (2024)
-
Chronic replication stress invokes mitochondria dysfunction via impaired parkin activity
Scientific Reports (2024)
-
Pathogenic mutations of human phosphorylation sites affect protein–protein interactions
Nature Communications (2024)
-
PINK1-PTEN axis promotes metastasis and chemoresistance in ovarian cancer via non-canonical pathway
Journal of Experimental & Clinical Cancer Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.