Abstract
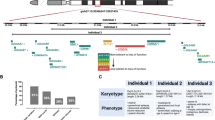
Trisomy 21 is the most frequent genetic cause of cognitive impairment. To assess the perturbations of gene expression in trisomy 21, and to eliminate the noise of genomic variability, we studied the transcriptome of fetal fibroblasts from a pair of monozygotic twins discordant for trisomy 21. Here we show that the differential expression between the twins is organized in domains along all chromosomes that are either upregulated or downregulated. These gene expression dysregulation domains (GEDDs) can be defined by the expression level of their gene content, and are well conserved in induced pluripotent stem cells derived from the twins’ fibroblasts. Comparison of the transcriptome of the Ts65Dn mouse model of Down’s syndrome and normal littermate mouse fibroblasts also showed GEDDs along the mouse chromosomes that were syntenic in human. The GEDDs correlate with the lamina-associated (LADs) and replication domains of mammalian cells. The overall position of LADs was not altered in trisomic cells; however, the H3K4me3 profile of the trisomic fibroblasts was modified and accurately followed the GEDD pattern. These results indicate that the nuclear compartments of trisomic cells undergo modifications of the chromatin environment influencing the overall transcriptome, and that GEDDs may therefore contribute to some trisomy 21 phenotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
02 December 2015
Nature 508, 345–350 (2014); doi:10.1038/nature13200 Owing to a labelling error in the input files, one of the two replicate data sets used for Fig. 5d and e and Supplementary Fig. 6d of this Article was incorrect. We have now repeated the analysis with a correct, independent replicate experiment. This confirms our previous conclusion that there are no detectable differences in nuclear lamina interactions between the normal and trisomy 21 twin cells.
References
Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A. & Deutsch, S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nature Rev. Genet. 5, 725–738 (2004)
Korenberg, J. R. et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl Acad. Sci. USA 91, 4997–5001 (1994)
Pritchard, M. A. & Kola, I. The “gene dosage effect” hypothesis versus the “amplified developmental instability” hypothesis in Down syndrome. J. Neural Transm. Suppl. 57, 293–303 (1999)
Lyle, R. et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 17, 454–466 (2009)
Shapiro, B. L. Down syndrome–a disruption of homeostasis. Am. J. Med. Genet. 14, 241–269 (1983)
Letourneau, A. & Antonarakis, S. E. Genomic determinants in the phenotypic variability of Down syndrome. Prog. Brain Res. 197, 15–28 (2012)
Davisson, M. T. et al. Segmental trisomy as a mouse model for Down syndrome. Prog. Clin. Biol. Res. 384, 117–133 (1993)
Reeves, R. H. et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nature Genet. 11, 177–184 (1995)
Chrast, R. et al. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9, 1853–1864 (2000)
Ahn, K. J. et al. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 22, 463–472 (2006)
Costa, V. et al. Massive-scale RNA-Seq analysis of non ribosomal transcriptome in human trisomy 21. PLoS ONE 6, e18493 (2011)
Esposito, G. et al. Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum. Mol. Genet. 17, 440–457 (2008)
Lockstone, H. E. et al. Gene expression profiling in the adult Down syndrome brain. Genomics 90, 647–660 (2007)
Sommer, C. A., Pavarino-Bertelli, E. C., Goloni-Bertollo, E. M. & Henrique-Silva, F. Identification of dysregulated genes in lymphocytes from children with Down syndrome. Genome 51, 19–29 (2008)
Prandini, P. et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am. J. Hum. Genet. 81, 252–263 (2007)
Deutsch, S. et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 14, 3741–3749 (2005)
Dahoun, S. et al. Monozygotic twins discordant for trisomy 21 and maternal 21q inheritance: a complex series of events. Am. J. Med. Genet. A. 146A, 2086–2093 (2008)
Marco-Sola, S., Sammeth, M., Guigo, R. & Ribeca, P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nature Methods 9, 1185–1188 (2012)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Ram, G. & Chinen, J. Infections and immunodeficiency in Down syndrome. Clin. Exp. Immunol. 164, 9–16 (2011)
Caron, H. et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291, 1289–1292 (2001)
Gierman, H. J. et al. Domain-wide regulation of gene expression in the human genome. Genome Res. 17, 1286–1295 (2007)
Hibaoui, Y. et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 6, 259–277 (2014)
Duchon, A. et al. Identification of the translocation breakpoints in the Ts65Dn and Ts1Cje mouse lines: relevance for modeling Down syndrome. Mamm. Genome 22, 674–684 (2011)
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013)
Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008)
Peric-Hupkes, D. et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010)
Zullo, J. M. et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012)
Akhtar, A. & Gasser, S. M. The nuclear envelope and transcriptional control. Nature Rev. Genet. 8, 507–517 (2007)
Vogel, M. J., Peric-Hupkes, D. & van Steensel, B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nature Protocols 2, 1467–1478 (2007)
Hansen, R. S. et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–144 (2010)
Gilbert, D. M. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14, 377–383 (2002)
Hiratani, I. et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 (2008)
Pope, B. D. et al. Replication-timing boundaries facilitate cell-type and species-specific regulation of a rearranged human chromosome in mouse. Hum. Mol. Genet. 21, 4162–4170 (2012)
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols 6, 468–481 (2011)
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007)
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
Singh, M. P., Wijeratne, S. S. & Zempleni, J. Biotinylation of lysine 16 in histone H4 contributes toward nucleosome condensation. Arch. Biochem. Biophys. 529, 105–111 (2013)
Pestinger, V., Wijeratne, S. S., Rodriguez-Melendez, R. & Zempleni, J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J. Nutr. Biochem. 22, 328–333 (2011)
Narang, M. A., Dumas, R., Ayer, L. M. & Gravel, R. A. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum. Mol. Genet. 13, 15–23 (2004)
Postnikov, Y. & Bustin, M. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta 1799, 62–68 (2010)
Zhu, N. & Hansen, U. Transcriptional regulation by HMGN proteins. Biochim. Biophys. Acta 1799, 74–79 (2010)
Canzonetta, C. et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 83, 388–400 (2008)
Huang, H., Rambaldi, I., Daniels, E. & Featherstone, M. Expression of the Wdr9 gene and protein products during mouse development. Dev. Dyn. 227, 608–614 (2003)
Bakshi, R. et al. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell. Physiol. 225, 569–576 (2010)
Contestabile, A. et al. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus 17, 665–678 (2007)
Williams, B. R. et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008)
Voss, T. C. & Hager, G. L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nature Rev. Genet. 15, 69–81 (2014)
Dimas, A. S. et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 (2009)
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007)
Grad, I. et al. NANOG priming before full reprogramming may generate germ cell tumours. Eur. Cells Mat. 22, 258–274 (2011)
Knowles, D. G., Roder, M., Merkel, A. & Guigo, R. Grape RNA-Seq analysis pipeline environment. Bioinformatics 29, 614–621 (2013)
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011)
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012)
John, S. et al. Genome-scale mapping of DNase I hypersensitivity. Curr. Protoc. Mol. Biol. Ch. 27, Unit 21 27. (2013)
Acknowledgements
We thank the Swiss National Science Foundation (SNF-144082), the European Research Council (ERC-249968), AnEUploidy and BluePrint EU grants, the Lejeune, and ChildCare foundations for supporting the S.E.A. laboratory. The laboratories of R.G. were supported by Spanish MICINN (BIO2011-26205) and ERC-294653, B.v.S. by NWO-ALW-VICI, Y.He. by CNRS, INSERM, University of Strasbourg and ANR-10-INBS-07, J.A.S. by NIH U54HG007010, and A.F. by Genico and Ernest Boninchi foundation. We thank S. Dahoun and J. L. Blouin for the discordant twins sample collection.
Author information
Authors and Affiliations
Contributions
The project was coordinated by S.E.A. A.L. coordinated/undertook the main laboratory work. F.A.S. coordinated/undertook the main bioinformatics/statistical analyses. X.B. performed ChIP-seq experiments. M.R.S. performed DNA methylation, A.L., F.A.S., M.G., R.G. and D.G. processed NGS data. J.K. and B.v.S. performed DamID experiments. C.C. and Y.He. maintained the mouse colony and contributed mouse samples. R.T., R.S.S. and J.A.S. performed DNase experiments; Y.Hi. and A.F. derived the iPS cells; and K.P., D.R. R.G. and E.M. performed additional statistical analyses. E.F., M.G., C.G., A.V., M.G., L.F., C.B. and S.D. assisted with wet lab experiments and contributed to performing NGS experiments. The main findings were interpreted by S.E.A., A.L. and F.A.S., who also wrote the manuscript. All authors made comments on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-12. (PDF 8032 kb)
Supplementary Table 1
Differential expression analysis results as given by EdgeR. For each gene, the table gives gene coordinates, Ensembl gene ID, gene category, gene symbol, log2 gene expression fold change (T1DS/T2N, all replicates), log2 counts-per-million (logCPM), p-value and false discovery rate (FDR). (XLSX 4151 kb)
Supplementary Table 2
List of GEDDs identified in the twins’ fibroblasts (Rep0). Table gives the domain ID, the names and coordinates of the first genes at the left and right borders of each GEDD, the number of genes included in the domain and the median log2 fold change within the domain. GEDDs were obtained with a smoothing bandwidth of 3%. (XLSX 38 kb)
Rights and permissions
About this article
Cite this article
Letourneau, A., Santoni, F., Bonilla, X. et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508, 345–350 (2014). https://doi.org/10.1038/nature13200
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13200
This article is cited by
-
Transcriptomic analysis of stem cells from chorionic villi uncovers the impact of chromosomes 2, 6 and 22 in the clinical manifestations of Down syndrome
Stem Cell Research & Therapy (2023)
-
Epigenomic signatures reveal mechanistic clues and predictive markers for autism spectrum disorder
Molecular Psychiatry (2023)
-
Consequences of gaining an extra chromosome
Chromosome Research (2023)
-
APP and DYRK1A regulate axonal and synaptic vesicle protein networks and mediate Alzheimer’s pathology in trisomy 21 neurons
Molecular Psychiatry (2022)
-
Differential microRNA expression profile in blood of children with Down syndrome suggests a role in immunological dysfunction
Human Cell (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.