Abstract
Tripartite Tc toxin complexes of bacterial pathogens perforate the host membrane and translocate toxic enzymes into the host cell, including in humans. The underlying mechanism is complex but poorly understood. Here we report the first, to our knowledge, high-resolution structures of a TcA subunit in its prepore and pore state and of a complete 1.7 megadalton Tc complex. The structures reveal that, in addition to a translocation channel, TcA forms four receptor-binding sites and a neuraminidase-like region, which are important for its host specificity. pH-induced opening of the shell releases an entropic spring that drives the injection of the TcA channel into the membrane. Binding of TcB/TcC to TcA opens a gate formed by a six-bladed β-propeller and results in a continuous protein translocation channel, whose architecture and properties suggest a novel mode of protein unfolding and translocation. Our results allow us to understand key steps of infections involving Tc toxins at the molecular level.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The coordinates for the EM structures have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-2551 and EMD-2552. Coordinates of TcdB2–TccC3 and TcdA1 have been deposited in the Protein Data Bank under accession numbers 4O9X and 4O9Y, respectively.
References
Waterfield, N. R., Bowen, D. J., Fetherston, J. D., Perry, R. D. & ffrench-Constant, R. H. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 9, 185–191 (2001)
ffrench-Constant, R. H. & Bowen, D. J. Novel insecticidal toxins from nematode-symbiotic bacteria. Cell. Mol. Life Sci. 57, 828–833 (2000)
Hares, M. C. et al. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology 154, 3503–3517 (2008)
Sheets, J. J. et al. Insecticidal toxin complex proteins from Xenorhabdus nematophilus: structure and pore formation. J. Biol. Chem. 286, 22742–22749 (2011)
Gatsogiannis, C. et al. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature 495, 520–523 (2013)
Bravo, A. & Soberón, M. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 26, 573–579 (2008)
ffrench-Constant, R. H., Eleftherianos, I. & Reynolds, S. E. A nematode symbiont sheds light on invertebrate immunity. Trends Parasitol. 23, 514–517 (2007)
Lang, A. E. et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 327, 1139–1142 (2010)
Busby, J. N., Panjikar, S., Landsberg, M. J., Hurst, M. R. H. & Lott, J. S. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501, 547–550 (2013)
Landsberg, M. J. et al. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc. Natl Acad. Sci. USA 108, 20544–20549 (2011)
Lee, S. C. et al. Structural characterisation of the insecticidal toxin XptA1, reveals a 1.15 MDa tetramer with a cage-like structure. J. Mol. Biol. 366, 1558–1568 (2007)
Lesieur, C., Vécsey-Semjén, B., Abrami, L., Fivaz, M. & Gisou van der Goot, F. Membrane insertion: the strategies of toxins. Mol. Membr. Biol. 14, 45–64 (1997)
Murphy, J. R. Mechanism of diphtheria toxin catalytic domain delivery to the eukaryotic cell cytosol and the cellular factors that directly participate in the process. Toxins 3, 294–308 (2011)
Young, J. A. T. & Collier, R. J. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 76, 243–265 (2007)
Varghese, J. N., Laver, W. G. & Colman, P. M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303, 35–40 (1983)
Choe, S. et al. The crystal structure of diphtheria toxin. Nature 357, 216–222 (1992)
Petosa, C., Collier, R. J., Klimpel, K. R., Leppla, S. H. & Liddington, R. C. Crystal structure of the anthrax toxin protective antigen. Nature 385, 833–838 (1997)
Li, Z., Strunk, J. J., Lamken, P., Piehler, J. & Walz, T. The EM structure of a type I interferon–receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 377, 715–724 (2008)
Humphries, M. J. Integrin structure. Biochem. Soc. Trans. 28, 311–339 (2000)
Lang, A. E., Konukiewitz, J., Aktories, K. & Benz, R. TcdA1 of Photorhabdus luminescens: electrophysiological analysis of pore formation and effector binding. Biophys. J. 105, 376–384 (2013)
Blaustein, R. O., Koehler, T. M., Collier, R. J. & Finkelstein, A. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc. Natl Acad. Sci. USA 86, 2209–2213 (1989)
Brown, M. J., Thoren, K. L. & Krantz, B. A. Charge requirements for proton gradient-driven translocation of anthrax toxin. J. Biol. Chem. 286, 23189–23199 (2011)
Feld, G. K., Brown, M. J. & Krantz, B. A. Ratcheting up protein translocation with anthrax toxin. Protein Sci. 21, 606–624 (2012)
Krantz, B. A. et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science 309, 777–781 (2005)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Berendsen, H. & van der Spoel, D. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. R. in Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography Vol. 33, 22–24 (European Science Foundation, 1997)
French, S. & Wilson, K. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Baker, D., Bystroff, C., Fletterick, R. J. & Agard, D. A. PRISM: topologically constrained phased refinement for macromolecular crystallography. Acta Crystallogr. D 49, 429–439 (1993)
Jones, T. A. A graphics model building and refinement system for macromolecules. J. Appl. Crystallogr. 11, 268–272 (1978)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Ludtke, S. J. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673, 157–173 (2010)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Raunser, S. et al. Rubisco in complex with Rubisco large subunit methyltransferase. Proc. Natl Acad. Sci. USA 106, 3160–3165 (2009)
Fernández, J. J., Luque, D., Castón, J. R. & Carrascosa, J. L. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 164, 170–175 (2008)
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013)
Harpaz, Y., Gerstein, M. & Chothia, C. Volume changes on protein folding. Structure 2, 641–649 (1994)
Wang, Z. & Schröder, G. F. Real-space refinement with DireX: from global fitting to side-chain improvements. Biopolymers 97, 687–697 (2012)
Hayward, S. & Lee, R. A. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graph. Model. 21, 181–183 (2002)
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Laskowski, R. A., Watson, J. D. & Thornton, J. M. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 33, W89–W93 (2005)
Pérez, A., Marchán, I., Svozil, D. & Sponer, J. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. (2007)
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Pellegrini-Calace, M., Maiwald, T. & Thornton, J. M. PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLOS Comput. Biol. 5, e1000440 (2009)
Jo, S., Lim, J. B., Klauda, J. B. & Im, W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009)
Walshaw, J. & Woolfson, D. N. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 307, 1427–1450 (2001)
Lemkul, J. A. & Bevan, D. R. Assessing the stability of Alzheimer’s amyloid protofibrils using molecular dynamics. J. Phys. Chem. B 114, 1652–1660 (2010)
Jones, D. T. et al. Prediction of novel and analogous folds using fragment assembly and fold recognition. Proteins 61 (suppl. 7). 143–151 (2005)
Kumar, S., Rosenberg, J. M., Bouzida, D., Swendsen, R. H. & Kollman, P. A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992)
Sotomayor, M. & Schulten, K. Single-molecule experiments in vitro and in silico. Science 316, 1144–1148 (2007)
Erickson, H. P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl Acad. Sci. USA 91, 10114–10118 (1994)
Flory, P. J. Theory of elastic mechanisms in fibrous proteins. J. Am. Chem. Soc. 78, 5222–5235 (1956)
Zhang, B., Xu, G. & Evans, J. S. A kinetic molecular model of the reversible unfolding and refolding of titin under force extension. Biophys. J. 77, 1306–1315 (1999)
Noy, A. Handbook of Molecular Force Spectroscopy (Springer, 2007)
Koenig, T. et al. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J. Proteome Res. 7, 3708–3717 (2008)
Zhang, Z. & Marshall, A. G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 9, 225–233 (1998)
Acknowledgements
We are grateful to R. S. Goody for continuous support and for useful comments on the manuscript. We thank V. Pfaumann for cloning, C. Schwan and S. Bergbrede for technical support and the Raunser laboratory for selecting particles. We thank the staff of beamline X10SA at the Paul Scherrer Institute, the X-ray communities at the Max Planck Institute (MPI) Dortmund and the MPI Heidelberg for help with data collection. This work was supported by the Deutsche Forschungsgemeinschaft, grants RA 1781/1-1 (to S.R.) and AK 6/22-1 (to K.A.), and by the Max Planck Society (to S.R.).
Author information
Authors and Affiliations
Contributions
D.M. crystallized both protein complexes. D.M., R.G.E. and I.R.V. processed, refined and analysed X-ray data. C.G. and O.H. collected cryo-EM data; C.G. processed, refined and analysed cryo-EM data; A.E.L. purified proteins and performed mutational studies; I.R.V. calculated molecular dynamics simulations; K.A. and S.R. designed the study. D.M., C.G., R.G.E. and A.E.L. prepared figures. S.R. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Topology diagram and structural homologues of selected TcA domains.
Receptor-binding domains A to D, neuraminidase-like domain and TcB-binding domain of TcA are coloured according to the colour code used in Fig. 1. Two closest structural homologues (Protein Data Bank (PDB) accession and corresponding root mean squared deviation (r.m.s.d.)) were aligned to the query structure.
Extended Data Figure 2 Biophysical properties of the TcA channel and the interface between two shell subunits in the prepore and pore state of TcA.
a–c, Top and side views on the surface and in the lumen of the channel, demonstrating the conservation of residues (a), the surface hydrophobic potential (b) and the surface electrostatic Coulomb potential at pH 7.5 (c). Conserved residues are depicted in purplish-red, hydrophobic patches are shown in orange, positive-charge and negative-charge density are coloured in blue and red, respectively. d, The putative transmembrane region (yellow) of the TcA prepore channel (amino acids 2104–2190) was integrated into a homogeneous dioleoyl-phosphatidylcholine lipid bilayer (115 × 115 × 40 Å) using molecular dynamics simulation. Spheres of D2186–2189 and E2104/2105 are highlighted in blue and red, respectively. e, Residues of the large interface between two shell subunits are depicted in yellow and green on the crystal structure of the TcA prepore state (left) and flipped open to demonstrate the complementary charges at the surface (electrostatic Coulomb potential at pH 7.5) (right). f, Same as in e, but for the TcA pore state.
Extended Data Figure 3 Cryo-EM of the TcA pore complex and the holotoxin complex PTC3 (TcA/TcB–TcC).
a, Representative digital micrograph area of vitrified TcA pore complexes. b, Corresponding representative class averages, each containing 300–500 single particles. Scale bars, 20 nm. c, Fourier shell correlation (FSC). The 0.5 FSC cut-off criterion indicates that the cryo-EM map has an average resolution of 9 Å. d, Surface renderings of the TcA-pore reconstruction in top (left) and side (right) view, coloured according to localized resolution. e, Fitting of the TcA prepore crystal structure in the electron density of the TcA pore. f, Fitting of the present cryo-EM reconstruction of the TcA pore complex (shown in red) in our previous low-resolution cryo-EM structure of the TcA pore complex reconstituted in liposomes (Electron Microscopy Data Bank accession EMD-2298) (grey). The arrow indicates that the transmembrane part of the channel is not resolved in the present structure of the pore. g, h, Top and side view of the molecular model of the TcA pore, obtained by flexible fitting in the cryo-EM density. The different domains are coloured according to the colour code used in Fig. 1. Parts of the model that were not resolved in the cryo-EM density map (receptor-binding domain A, transmembrane part of the central channel) are indicated by black arrows. i, A typical digital micrograph area of PTC3 holotoxin complexes in vitrified ice. Scale bar, 20 nm. j, Characteristic reference-free class averages of PTC3 complexes, each containing 300–400 single particles. Scale bar, 10 nm. k, The 0.5 FSC cut-off criterion indicates that the cryo-EM maps of PTC3 (TcA/TcB–TcC, C1 symmetry—that is, no symmetry) and TcA (TcB–TcC was masked during image processing, C5 symmetry) have an average resolution of 9.1 Å and 7.4 Å, respectively. l, Surface and cross-sections of PTC3 and TcA coloured according to the local resolution. For better clarity the respective maps are filtered to their average resolutions. See Methods for more detail.
Extended Data Figure 4 Flexible regions and hinges of TcA.
a, Structure of the TcA prepore protomer, with the different domains coloured according to the colour code used in Fig. 1. Four groups of domains were identified as ‘quasi-rigid bodies’ during motion analysis and are highlighted in different colours (the large lobe of the α-helical domain, red; receptor-binding domain D and the neuraminidase-like domain, yellow; receptor-binding domain B and receptor-binding domain C, cyan; the pore-forming domain and the TcB-binding domain, grey). b, Structure of the TcA prepore protomer, with the four bending regions as identified by the domain motion analysis highlighted in magenta. c–f, Superimposition of the quasi-rigid bodies of TcA in the prepore (coloured) and pore state (white) for the four different pairs of covalently linked quasi-rigid bodies, as highlighted in a. Details of the interdomain motion are given for each pair.
Extended Data Figure 5 The hyperconserved region of TcC, the interior of the TcB–TcC cocoon and the interaction surface between TcB and TcC.
a, Ribbon representation of the hyperconserved region of TcC (green). Aspartic acid residues at the intrinsic aspartyl protease site are coloured in dark blue. D651 and D674 built the actual catalytic dyad, whereas D657 and D676 are not directly involved in proteolysis. b, Topology diagram of the hyperconserved region of TcC. Positions of the two highly conserved catalytic aspartate residues (D651 and D674) are marked with an asterisk. c, d, Amino acid sequences of the hyperconserved region of TcC and 18 homologues were aligned. The conservation score corresponds to the percentage of conserved amino acid residues and is coloured from cyan for no conservation to magenta for complete conservation. The two conserved catalytic aspartic acid residues (D651 and D674) of the intrinsic aspartyl protease domain are marked with an asterisk. e, f, Hyperconserved region of TcC (yellow) (e) as well as the TcB N-terminal region (dark blue) and TcA-binding domain (light blue) (f). The crystal structure of YenB and YenC2 from Yersinia entomophaga (coloured in red) (PDB accession 4IGL) are overlaid for comparison. The depicted domains match each other with an overall r.m.s.d. of 0.88 Å. g, Conservation of amino acid residues at the interface between TcB and TcC. Conservation score corresponds to the percentage of conserved amino acid residues and is coloured from cyan for no (0%) conservation to magenta for complete (100%) conservation. h–k, Sliced views displaying the surface hydrophobic potential (h, j) and the surface electrostatic Coulomb potential at pH 7.5 (i, k) of the inner surface of TcB–TcC. Hydrophobic patches are shown in orange, positively and negatively charged surfaces are coloured in blue and red, respectively.
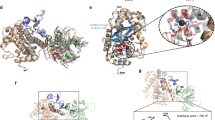
Extended Data Figure 6 TcA-binding domain of TcB and the binding interface between TcA and TcB.
a, Ribbon presentation of TcA-binding domain and its homologues. Two closest structural homologues (PDB accession and corresponding r.m.s.d.) were aligned to the query structure. b, Surface hydrophobicity of the four symmetric blades of the TcA-binding domain. Note the hydrophobic surface in the central passage of the β-propeller of the TcA-binding domain through which the ADP-ribosyltransferase passes before it enters the TcA translocation channel. c, d, Ribbon presentation (c) and topology diagram (d) of the TcA-binding domain (amino acids 374–693) of TcB. Blades 1 to 4 of the six-bladed β-propeller are coloured from dark to light blue whereas distorted blades 5 and 6 are coloured in red and orange, respectively. e, Sequence alignment of the six β-propeller blade regions. The two glycines of the DxxGxGxxxL(W)V(I) repeat (blue) are highlighted in the sequence with an asterisk and displayed in spheres in the structure depicted in a. Note the different orientation of both glycines in distorted blade 6 compared with the glycines in the other blades. f, Specific interaction between TcA (TcB-binding domain) depicted as surface representation and TcB–TcC (β-propeller of the TcA-binding domain) depicted as ribbon representation. The β-hairpin that closes the central passage of the β-propeller before its interaction with TcA is depicted in yellow. Positively and negatively charged surfaces are coloured in blue and red, respectively. Hydrophobic patches are depicted in orange. g, The electron density of the TcB–β-propeller in the holotoxin complex has pseudo-five-fold symmetry. h, Rigid-body fit of the TcA-binding domain into the corresponding density. Arrows indicate putative conformational changes that are necessary to open the central passage of the β-propeller of the TcA-binding domain. The β-hairpin that closes the central passage of the β-propeller before its interaction with TcdA1 is depicted in red.
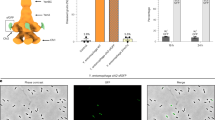
Extended Data Figure 7 Cleavage of the C-terminal ADP-ribosyltransferase domain and in vivo intoxication with wild-type and mutant TcC.
a, Two micrograms of recombinant wild-type (WT) and mutant TcC (TccC3), fused to TcB, were loaded onto a gradient SDS gel (3–12% SDS) and stained with Coomassie blue (top). Wild-type and mutant TcB–TcC were subjected to SDS–PAGE (12.5% SDS) and the C-terminal hvr domain was detected by immunoblotting (bottom). Note, in uncleaved TcB–TcC, the hvr domain is detected at ∼280 kDa. The data are representative of four repetitions. b, ADP-ribosylation of actin induces actin aggregation that can be detected by fluorescence microscopy. Intoxication of HeLa cells with wild-type and mutant TcC. All samples were treated for 2 h with TcA (2 µg ml−1) and the indicated wild-type or mutant TcB–TcC complexes (1 µg ml−1). Untreated cells were used as controls (con). Then, cells were fixed and stained with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated phalloidin and 4′,6-diamidino-2-phenylindole (DAPI). For each construct, ten different cells were imaged. The experiment was replicated three times. Scale bar, 10 µm. c, SDS–PAGE of 15–20 TcB–TcC crystals. The gel represents one out of four crystals that were screened, yielding the same result. Protein bands marked with an arrow were digested with trypsin before high-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis. HPLC-MS analysis clearly identified this band as the ADP-ribosyltransferase domain, showing that this domain is located in our crystals but cleaved off from the rest of the protein. d, Electronspray ionization (ESI)-MS measurements of TcB–TcC showed that the ADP-ribosyltransferase domain is trapped inside the TcB–TcC cocoon, as the measured molecular weight of the sample corresponds to the full-length complex, including the ADP-ribosyltransferase domain.
Extended Data Figure 8 Mechanism of action of Tc toxin complexes in insect hosts.
a, P. luminescens secretes Tc toxin complexes that attack insect midgut cells. We propose that this can happen from the haemocoel, that is, the basolateral membrane, or the midgut lumen, that is, the apical membrane. The C-terminal hypervariable region of TccC3 or TccC5, which are both ADP-ribosyltransferases, is trapped in a closed cocoon, formed by the large β-sheet structures of TcdB2–TccC3 or TcdB2–TccC5, respectively. It is probably unfolded inside the hostile environment of the cocoon and autoproteolytically cleaved by the intrinsic aspartate protease activity of TcC. b, With the assembly of the holotoxin, that is, complex formation between TcB–TcC and TcA, the closed gate inside the connecting β-propeller opens owing to an induced fit mechanism, resulting in a long continuous channel. The ADP-ribosyltransferase penetrates into the channel of TcA. However, the channel is blocked until TcA permeates the membrane. c, The PTC3 holotoxin complex binds with its receptor-binding domains to receptors on the surface of host cells. d, This triggers its endocytosis on the basolateral membrane. e, High pH values in the midgut lumen or low pH values in endosomes open the electrostatic lock formed by the neuraminidase-like domain of TcA, which results in an opening of the shell. The linker between the shell and channel of TcA, which acts as an entropic spring, is released and drives the syringe-like injection of the TcA translocation channel into the membrane. f, The ADP-ribosyltransferase is translocated through the TcA pore into the host cytoplasm. g, The ADP-ribosyltransferase corresponding to the C-terminal hypervariable region of TccC3 modifies G-actin such that it cannot bind to β-thymosin-4 anymore. The ADP-ribosyltransferase corresponding to the C-terminal hypervariable region of TccC5 fixes RhoA in its active state. Both modifications result in uncontrolled actin polymerization, collapse of the cytoskeleton and ultimately to cell death.
Extended Data Figure 9 Representative model-based and experimentally phased electron density maps of TcdA1 and TcdB2–TccC3.
Visualization of the experimentally phased 4 Å and 2.35 Å resolution electron density maps (purple) of the TcB-binding and α-helical pore-forming domain of TcdA1 (top left) and the β-sheet-rich cocoon region of TcdB2–TccC3 (top right) contoured at 1.5σ, respectively. The 2Fobs−Fcalc Fourier maps (blue) of TcdA1 at 3.5 Å resolution (bottom left) show the interaction interface between two protomers, whereas the Fourier maps of TcdB2–TccC3 at 2.5 Å resolution (bottom right) represent distinct amino acid side chains in the β-sheet cocoon contoured at 1.5σ, respectively.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion including a detailed description of the TcdB2-TccC3 structure and mechanism of action of Tc toxins, Supplementary references and Supplementary Table 1. (PDF 218 kb)
Architecture of Tc toxins
First, the video shows the crystal structure of a TcA protomer in its prepore state with the different domains coloured and labelled according to Figure 1 and subsequently its pentameric assembly. Then, the crystal structure of TcB-TcC is shown (Figure 4). Subsequently, the video shows a rigid body fit of both crystal structures (TcA-pentamer, TcB/TcC) into the cryo-EM structure of the holotoxin complex PTC3. The video focuses then on the central channel, depicting the fitted α-helices and the electrostatic lock (Figure 2). In the end, the molecular model of the holotoxin is shown (Figure 4). (MP4 29128 kb)
TcA membrane insertion driven by an entropic-spring injection mechanism
The side view of a morph between the prepore and the pore model of the TcA protomer and pentamer. The different domains are coloured according to the colour code used in Figure 1. A grey bar indicates the membrane bilayer. (MP4 4637 kb)
Binding of TcB-TcC to TcA opens a molecular gate
This video shows cryo-EM structures of the TcB-TcC and TcA complex and their assembly. It then zooms in and shows that the ß-propeller gate is open, so that the TcB-TcC cocoon opens towards the lumen of the TcA translocation pore. Finally the interface between the TcA-binding domain and the TcB-binding domain is shown. (MOV 13018 kb)
Rights and permissions
About this article
Cite this article
Meusch, D., Gatsogiannis, C., Efremov, R. et al. Mechanism of Tc toxin action revealed in molecular detail. Nature 508, 61–65 (2014). https://doi.org/10.1038/nature13015
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13015
This article is cited by
-
Stepwise assembly and release of Tc toxins from Yersinia entomophaga
Nature Microbiology (2024)
-
Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations
Nature Microbiology (2024)
-
CRISPR screens in Drosophila cells identify Vsg as a Tc toxin receptor
Nature (2022)
-
Development of a cost-effective medium for Photorhabdus temperata bioinsecticide production from wastewater and exploration of performance kinetic
Scientific Reports (2021)
-
The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: a review
Parasites & Vectors (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.