Abstract
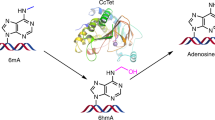
Cytosine residues in mammalian DNA occur in five forms: cytosine (C), 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). The ten-eleven translocation (Tet) dioxygenases convert 5mC to 5hmC, 5fC and 5caC in three consecutive, Fe(ii)- and α-ketoglutarate-dependent oxidation reactions1,2,3,4. The Tet family of dioxygenases is widely distributed across the tree of life5, including in the heterolobosean amoeboflagellate Naegleria gruberi. The genome of Naegleria6 encodes homologues of mammalian DNA methyltransferase and Tet proteins7. Here we study biochemically and structurally one of the Naegleria Tet-like proteins (NgTet1), which shares significant sequence conservation (approximately 14% identity or 39% similarity) with mammalian Tet1. Like mammalian Tet proteins, NgTet1 acts on 5mC and generates 5hmC, 5fC and 5caC. The crystal structure of NgTet1 in complex with DNA containing a 5mCpG site revealed that NgTet1 uses a base-flipping mechanism to access 5mC. The DNA is contacted from the minor groove and bent towards the major groove. The flipped 5mC is positioned in the active-site pocket with planar stacking contacts, Watson–Crick polar hydrogen bonds and van der Waals interactions specific for 5mC. The sequence conservation between NgTet1 and mammalian Tet1, including residues involved in structural integrity and functional significance, suggests structural conservation across phyla.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010)
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011)
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011)
Iyer, L. M., Zhang, D., Maxwell Burroughs, A. & Aravind, L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 41, 7635–7655 (2013)
Fritz-Laylin, L. K. et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642 (2010)
Iyer, L. M., Abhiman, S. & Aravind, L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 101, 25–104 (2011)
Aik, W., McDonough, M. A., Thalhammer, A., Chowdhury, R. & Schofield, C. J. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struct. Biol. 22, 691–700 (2012)
McDonough, M. A., Loenarz, C., Chowdhury, R., Clifton, I. J. & Schofield, C. J. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 20, 659–672 (2010)
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002)
Yang, C. G. et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 (2008)
Yi, C. et al. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature 468, 330–333 (2010)
Klimasauskas, S., Kumar, S., Roberts, R. J. & Cheng, X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 (1994)
Slupphaug, G. et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 (1996)
Roberts, R. J. & Cheng, X. Base flipping. Annu. Rev. Biochem. 67, 181–198 (1998)
Horton, J. R. et al. Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase. Nucleic Acids Res. 32, 3877–3886 (2004)
Werner, R. M. et al. Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase. Biochemistry 39, 12585–12594 (2000)
Sun, Z. et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 3, 567–576 (2013)
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012)
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011)
Upadhyay, A. K., Horton, J. R., Zhang, X. & Cheng, X. Coordinated methyl-lysine erasure: structural and functional linkage of a Jumonji demethylase domain and a reader domain. Curr. Opin. Struct. Biol. 21, 750–760 (2011)
Fang, R. et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 49, 558–570 (2013)
Szulwach, K. E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neurosci. 14, 1607–1616 (2011)
Inoue, A., Shen, L., Dai, Q., He, C. & Zhang, Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21, 1670–1676 (2011)
Nestor, C. E. et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 22, 467–477 (2012)
Münzel, M. et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. 49, 5375–5377 (2010)
Haffner, M. C. et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637 (2011)
Stroud, H., Feng, S., Morey Kinney, S., Pradhan, S. & Jacobsen, S. E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 12, R54 (2011)
Hashimoto, H., Hong, S., Bhagwat, A. S., Zhang, X. & Cheng, X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 40, 10203–10214 (2012)
Otwinowski, Z., Borek, D., Majewski, W. & Minor, W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003)
Fu, Z.-Q., Chrzes, J., Sheldrick, G. M., Rose, J. & Wang, B.-C. A parallel program using SHELXD for quick heavy-atom partial structural solution on high-performance computers. J. Appl. Cryst. 40, 387–390 (2007)
Fu, Z. Q., Rose, J. & Wang, B. C. SGXPro: a parallel workflow engine enabling optimization of program performance and automation of structure determination. Acta Crystallogr. D 61, 951–959 (2005)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Maiti, A. & Drohat, A. C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011)
Zhang, L., Yu, M. & He, C. Mouse Tet1 protein can oxidize 5mC to 5hmC and 5caC on single-stranded DNA. Acta Chimi. Sin. 70, 2123–2126 (2012)
Pfaffeneder, T. et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. 50, 7008–7012 (2011)
Fu, Y. & He, C. Nucleic acid modifications with epigenetic significance. Curr. Opin. Chem. Biol. 16, 516–524 (2012)
Elkins, J. M. et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278, 1802–1806 (2003)
Yu, B. et al. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439, 879–884 (2006)
Price, J. C., Barr, E. W., Tirupati, B., Bollinger, J. M., Jr & Krebs, C. The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: a high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from . Escherichia coli. Biochemistry 42, 7497–7508 (2003)
Acknowledgements
We thank R. J. Roberts who initiated this collaborative work, and participated both in the work and the writing of the manuscript. We thank J. R. Horton for critical comments and B. Baker for synthesizing the oligonucleotides. Y.Z. thanks C. Fulton for helpful discussions on N. gruberi biology. The Department of Biochemistry of Emory University School of Medicine supported the use of SER-CAT beamlines. This work was supported by grants from the National Institutes of Health GM049245 to X.C. (who is a Georgia Research Alliance Eminent Scholar) and GM095209 and GM105132 to Y.Z.
Author information
Authors and Affiliations
Contributions
H.H. performed antibody-based and TDG-based activity assays, crystallographic experiments and expression of mouse Tet1 in HEK293T cells. X.Z. made the overexpression construct in E. coli, developed (together with H.H.) assay conditions and performed NgTet1-8 sequence analysis. J.E.P. and L.S. performed kinetic assays using the LC–MS method and J.E.P. characterized the mutants. Z.-Q.F. performed crystallographic phasing calculations and generated an initial poly-alanine model. N.D. and I.R.C. developed the LC–MS method for detection of modified cytosine residues. X.Z., Y.Z. and X.C. organized and designed the scope of the study, and all were involved in analysing data and preparing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y.Z., J.E.P., L.S., N.D. and I.R.C. have filed a patent application based on NgTet1.
Extended data figures and tables
Extended Data Figure 1 Sequence alignment of Naegleria Tet-like dioxygenases (1 to 8).
a, Schematic representation of NgTet1–8. b, Secondary structural elements are indicated in green (helices), blue (the major sheet) and cyan (the minor sheet). Numbering above the sequences corresponds to NgTet1. White-on-red residues are invariant among the eight sequences examined, whereas black or grey-highlighted positions are conserved substitutions). Positions highlighted are responsible for various functions as indicated: t for structural turn, h for hydrophobic core, 5mC for 5mC binding, α or αKG for binding of αKG or NOG, P for DNA phosphate interaction, i for intra-molecular polar interaction (D268) and ? for side chain of K137 pointing to the DNA major groove (panel c). Sequences included are NgTet1 (XP_002667965.1), NgTet2 (XP_002682154.1), NgTet3 (XP_002668005.1), NgTet4 (XP_002676528.1), NgTet5 (XP_002668409.1), NgTet6 (XP_002674105.1), NgTet7 (XP_002668594.1), and NgTet8 (XP_002676954.1). However, the N-terminal sequences for NgTet3, 5, 7, 8 are extended in frame to include more conserved sequence elements until either a putative initiation methionine or the end of a sequence contig (that is, until a sequencing gap is encountered), therefore the exact N terminus is unknown. For NgTet6, the sequence of XP_002674105 (177 residues) is probably incomplete at the N terminus. Extending scaffold 42_31984–32508 to 32980 and allowing for a proper splicing junction results in a protein of 313 residues that shares 51% identity with NgTet1 across the whole protein except for the first 20 residues. Of the five NgTet proteins tested (NgTet1–5), two of them (NgTet1 and NgTet4) have 5mC dioxygenase activities. c, An invariant Lys 137 among the eight NgTet dioxygenases, located in the loop between helix α2 and strand β5, points to the major groove of DNA with the terminal ε-amino group approximately 4.3 Å away from the base 3′ to the target 5mCpG site. An exchange of a C:G to G:C pair at this position does not affect crystallization. The corresponding loop in AlkB is the long loop L3 (see Fig. 3b) that makes DNA backbone contacts. In mammalian Tet1, the Cys-rich region is predicted to insert within the corresponding loop L3 (see Fig. 4c).
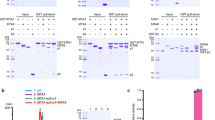
Extended Data Figure 2 Activity of NgTet1 on various DNA substrates.
a–c, The time courses (lanes 5–13) of the reactions using 32-bp DNA substrates containing 5mC (a), 5hmC (b) or 5caC (c). Lanes 1–4: antibody sensitivity against 10 pmol of control oligonucleotides and 2 fold serial dilutions. Lanes 5–13: the rate of conversion appears to be the fastest for the reaction of 5mC to 5hmC, and decreases with each subsequent reaction: 5mC to 5hmC > 5hmC to 5fC > 5fC to 5caC. d, Activities of NgTet1 (20 µM) on genomic DNA of Hela cells (2.5 µg). After 1 h reaction, 87% of the products are 5caC in gDNA with the remaining being 5fC and 5hmC. The percentages were estimated from integration of the peaks in LC–MS traces. The mean and standard error ( ± s.e.m.) were estimated from three repeated experiments. e, Human thymine DNA glycosylase (TDG) excises 5fC and 5caC (but not 5mC and 5hmC) when paired with a guanine in a CpG sequence (lanes 1–4)4,29,36. After NgTet1 reactions with DNA substrates containing 5mC, 5hmC or 5fC, in the presence of αKG, the product DNA containing 5fC and 5caC becomes a substrate for TDG (lanes 6, 8 and 10), but not with NOG (lanes 5 and 7), again demonstrating the production of 5fC and 5caC by NgTet1. f, Activities of NgTet1 on 56-bp double-stranded (ds) DNA-2 with methylation on both strands (M/M) or single strand (hemi-methylated either on top M/C or bottom C/M strand) or single-stranded (ss) DNA (reaction time 1 h and ± s.e.m. estimated from three repeats). We note that an in vitro activity of the mouse Tet1 catalytic domain on single-stranded DNA has also been observed37. g, LC–MS traces of a sample reaction mix on the hemi-methylated 5mCpG dsDNA-1 (top panel), reaction control with no enzyme (middle panel), and the standard deoxyribonucleoside mix (bottom panel). Arrows indicate peaks of 5mC, 5hmC, 5fC and 5caC. Identities of the peaks are confirmed by comparing the retention time with the standard as well as by mass spectrometry.
Extended Data Figure 3 Structure of NgTet1–DNA complex.
a, Schematic NgTet1–DNA interactions. b, The amino end of the 310-helix h3 interacts with the DNA backbone phosphate 3′ to the 5mCpG site. An arrow indicates the helical dipole. c, Unlike other DNA base flipping enzymes such as DNA methyltransferases13 and DNA repair glycosylases14, NgTet1 lacks a finger residue to occupy the space left by the everted 5mC. Instead, solvent molecules maintain the base stacking surrounding the flipped nucleotide. An ethylene glycol and a water molecule (behind ethylene glycol) occupy the space left by the everted 5mC. d, Superimposition of a normal intrahelical 5mC (coloured in grey) onto the flipped 5mC suggests a small rotation around the glycosidic bond. e, The simulated annealing omit electron density, contoured at 2.5σ above the mean, by omitting entire 14-bp DNA (approximately 21% of total content in the crystal). The density is shown for the length of the unit cell along the a axis (indicated by vertical grey lines). The bent DNA molecules mediate crystal packing contacts along the a axis by two-fold symmetry. The flipped 5mC is clearly visible in the active site (indicated by red circles). The broken density for the outer DNA bases in one end (as indicated by an arrow), in the absence of any protein contacts, correlates with higher crystallographic thermal B-factors (∼90 Å2) than that for the central DNA base pairs including 5mCpG (∼50 Å2) or those of the other end (∼67 Å2). f, g, Enlarged panels showing NgTet1 structure in two views.
Extended Data Figure 4 Modelling of 5hmC, 5fC and 5caC in the active site of NgTet1.
a, Superimposition of bases of 5hmC (cyan), 5fC (green), and 5caC (magenta) onto the flipped 5mC (yellow) in the NgTet1 active site. The closest residue to the 5-position modifications is Val 293 of strand β12 and Ala 212 of strand β6 (see panels c–e). The interaction involving hydrophobic Ala 212 and Val 293 might be the reason that the enzyme prefers 5mC (carrying a hydrophobic methyl group) over 5hmC (carrying a hydroxyl oxygen) or 5fC (carrying a carbonyl oxygen atom). b, The carboxylate group of 5caC (the final product of oxidation reaction by NgTet1) would be in the vicinity of the C1 carboxylate group of NOG (it would be succinate during the reaction cycle—see Extended Data Fig. 5), resulting in repulsion. c, Space filling model of 5hmC. The atoms are coloured with blue for nitrogen, red for oxygen, grey for carbon. The hydroxymethyl moiety of 5hmC is coloured in yellow (CH2) and its hydroxyl oxygen atom is in close contact with either the side chain of Val 293 (as shown) or Ala 212 (not shown). d, Space filling model of 5fC. The carbon atoms of 5fC are coloured either as green (ring carbon) or yellow (the formyl carbon). A study suggested the existence of a hydrated form of 5fC in DNA containing synthetic 5fC at a level of about 0.5%38. Because further oxidation of 5fC to 5caC would require the addition of water to the formyl group, the hydrated form of 5fC might be the real substrate during the oxidation of 5fC to 5caC39. Our structure may provide evidence in support of this hypothesis. A water molecule, held in place by Asn 214 and Tyr 141, might provide the water molecule needed for the formation of 5fC hydrate. e, Space filling model of 5caC. The carbon atoms of 5caC are coloured either as magenta (ring carbon) or yellow (the carboxylate carbon). The negatively charged carboxylate groups of 5caC and the carboxylate group of NOG would result in repulsion (right panel). f, We could model a water molecule with two alternative positions (left panel) or a dioxygen O2 molecule (right panel) as the sixth metal ligand as observed in the electron density 2Fo − Fc, contoured at 1σ above the mean. Previously, we studied a Jumonji PHF2–metal interaction (PDB 3PU8), where a water molecule was modelled as the sixth ligand. Comparing the two structures, we concluded that the density observed in NgTet1 active site is more than a water molecule and the density was best fit with either a water molecule with dual positions or an O2 molecule or a mixture of both. However, we do note that the observation of a dioxygen molecule needs to be confirmed independently by other methods.
Extended Data Figure 5 Proposed mechanism of 5mC oxidation by Fe(ii)- and αKG-dependent NgTet1.
We suggest an ordered binding of αKG (step 1) followed by DNA (step 2), DNA bending and base flipping by NgTet1. Like many base-flipping enzymes, NgTet1 might use a multi-step flipping pathway to distinguish substrate (5mC, 5hmC and 5fC) from non-substrate (unmodified C). The discrimination step could occur either before flipping when the C:G pair is intrahelical, during the flipping or after flipping where the nucleotide becomes extrahelical. The hydroxylation reaction is proposed to involve an Fe(iii)-superoxo intermediate which converts to a reactive Fe(iv)-oxo upon decarboxylation of αKG (step 3)42. The hydroxylated DNA is subsequently released (step 4), followed by exchange of succinate with αKG (step 5 and step 1) for next round of reaction. We do not know whether NgTet1 acts on DNA substrates distributively or processively (step 4) for the three consecutive, oxidation reactions that convert 5mC to 5caC. Metal ions Zn(ii), Mn(ii) or Co(ii) have been used to replace Fe(ii) in the studies of other dioxygenases, for example FIH40 and AlkB11,41; they occupy Fe(ii)-binding site but do not support catalysis. Like αKG, NOG (shown in the middle), initially used as an inhibitor in the study of FIH40, is ligated to Fe(ii) or Mn(ii) in a bidentate manner but does not support catalysis due to decreased susceptibility to attack by an Fe(iii)-superoxo. We used the combination of Mn(ii) and NOG in a very similar fashion as Zn(ii) and NOG used in the FIH study.
Extended Data Figure 6 Pairwise comparison of Naegleria NgTet1 and E. coli AlkB (panels a–c) or human ABH2 (panels d–f).
a, Structure-based sequence alignment of NgTet1 (PDB 4LT5) and AlkB (PDB 3O1M). NgTet1 has N-terminal as well as C-terminal additions. Secondary structural elements and residue numbering are indicated above (NgTet1) or below (AlkB) the sequences. Shared structural elements are coloured in green (helices), blue (the major sheet) and cyan (the minor sheet). White-on-red residues are invariant residues between the two, important for binding of metal ion and αKG, white-on-black are invariant for the hydrophobic core (h), structural turns (t) before or after β strands (glycine and proline residues), and intra-molecule interaction (see panel b). Grey-highlighted positions are conserved substitutions. The two proteins share 19 invariant residues that are important for metal ion coordination, αKG binding, hydrophobic packing and intramolecular interactions, as well as glycine and proline residues essential for structural turns before or after β strands. b, An invariant aspartate (Asp 268 in NgTet1 and Asp 174 in AlkB), located in strand β10, performs a network of stabilizing polar interactions with the main-chain amide nitrogen atoms immediately after strand β4 and β9. c, Superimposition of active sites of NgTet1 and AlkB indicate a co-variation of the binding site of the target base (5mC or 3mC) and the location of an arginine (Arg 224 of NgTet1 and Arg 210 of AlkB) that suggest conserved reaction chemistry and a conserved ion-pair interaction with the C1 carboxylate group of NOG of NgTet1 or αKG of AlkB. Arg 224 of NgTet1, located in the 310-helix h7, interacts with the C1 carboxylate group of NOG in a bidentate manner. Superimposition of NgTet1 and AlkB indicated that the flipped 3mC occupies the space of Arg 224 of NgTet1. Instead, AlkB uses Arg 210 of strand β12 of the major sheet, from the opposite direction of Arg 224 of NgTet1, to interact with the C1 carboxylate group or αKG. In NgTet1, the NOG molecule is involved in extensive interactions with the protein, including the carboxylate groups at C1 and C5 positions interacting with two arginine residues (Arg 224 of h7 and Arg 289 of β12), respectively, hydrophobic interactions with the side chains of Ile 225 of h7, Leu 240 of β7 and Leu 253 of β8 and polar interactions with the side chains of Asn 214 of β6 and Tyr 242 of β7 (see Fig. 2k, l). d, Structure-based sequence alignment of NgTet1 and hABH2. e, ABH2 (coloured in yellow; PDB 3BUC) has a hairpin loop insertion L3 between helix α2 and strand β5 and a 12-residue insertion between strands β8 and β9. NgTet1 (coloured in green) has the TET/JBP1-specific structural element (helices α5 and α6) and a C-terminal addition (helices α9 and α10). The two proteins share 28 invariant residues. f, The DNA molecules, bound with NgTet1 or hABH2, lie nearly perpendicular to each other relative to the proteins. We also note that the AlkB-DNA and ABH2-DNA complexes were captured by chemical cross-linking between an engineered mutant S129C, located in the AlkB-specific strand (coloured magenta in Fig. 3b) next to β11 as part of the minor sheet, and a disulphide-modified cytosine two nucleotides 3′ to the target base11,12.
Extended Data Figure 7 Pairwise comparison of Naegleria NgTet1 (coloured in green) and human ABH3 (magenta) (a), human FTO (cyan) (b), Chlamydomonas P4H (brown) (c) or human TYW5 (grey) (d).
Among them, NgTet1 has the TET/JBP1-specific structural element (helices α5 and α6). a, Like ABH2, ABH3 (PDB 2IUW) has a hairpin loop L3 insertion between helix α2 and strand β5 and a 13-residue insertion between strands β8 and β9. The two proteins share 30 invariant residues. b, FTO (PDB 3LFM) has a hairpin loop insertion between helix α2 and strand β5, a ∼30-residue insertion in the location corresponding to the helices α5 and α6 of NgTet1 and a 15-residue insertion between strands β7 and β8. The two proteins share 26 invariant residues. c, Chlamydomonas reinhardtii prolyl-4 hydroxylase type I (P4H, PDB 2JIJ) has insertions before strand β5, between strands β6 and β7, strands β8 and β9 and strands β10 and β11. The two proteins share 29 invariant residues. d. The tRNA wybutosine (yW)-synthesizing enzyme 5 (TYW5, PDB 3AL6) has large insertions between helix α2 and strand β5 and between strands β8 and β9, in addition to a C-terminal domain. The two proteins share 30 invariant residues.
Rights and permissions
About this article
Cite this article
Hashimoto, H., Pais, J., Zhang, X. et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 (2014). https://doi.org/10.1038/nature12905
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12905
This article is cited by
-
Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte
Nature (2018)
-
Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation
Nature Structural & Molecular Biology (2016)
-
Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism
Nature Communications (2016)
-
Structural insight into substrate preference for TET-mediated oxidation
Nature (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.