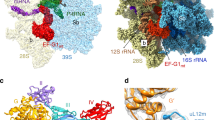
Abstract
Mitochondrial ribosomes synthesize a number of highly hydrophobic proteins encoded on the genome of mitochondria, the organelles in eukaryotic cells that are responsible for energy conversion by oxidative phosphorylation. The ribosomes in mammalian mitochondria have undergone massive structural changes throughout their evolution, including ribosomal RNA shortening and acquisition of mitochondria-specific ribosomal proteins. Here we present the three-dimensional structure of the 39S large subunit of the porcine mitochondrial ribosome determined by cryo-electron microscopy at 4.9 Å resolution. The structure, combined with data from chemical crosslinking and mass spectrometry experiments, reveals the unique features of the 39S subunit at near-atomic resolution and provides detailed insight into the architecture of the polypeptide exit site. This region of the mitochondrial ribosome has been considerably remodelled compared to its bacterial counterpart, providing a specialized platform for the synthesis and membrane insertion of the highly hydrophobic protein components of the respiratory chain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Protein Data Bank
Data deposits
The cryo-EM map of the 39S mitoribosomal subunit has been deposited in the Electron Microscopy Databank with accession code EMD-2490. The coordinates of the cryo-EM-based model of the 39S mitoribosomal subunit have been deposited in the Protein Data Bank under accession code 4CE4.
References
Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 14, 225–274 (1967)
Desmond, E., Brochier-Armanet, C., Forterre, P. & Gribaldo, S. On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Res. Microbiol. 162, 53–70 (2011)
Suzuki, T. et al. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 276, 21724–21736 (2001)
Koc, E. C. et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 276, 43958–43969 (2001)
Smits, P., Smeitink, J. A. M., van den Heuvel, L. P., Huynen, M. A. & Ettema, T. J. G. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 35, 4686–4703 (2007)
Attardi, G. & Ojala, D. Mitochondrial ribosome in HeLa cells. Nat. New Biol. 229, 133–136 (1971)
Ott, M. & Herrmann, J. M. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim. Biophys. Acta 1803, 767–775 (2010)
Ott, M. et al. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25, 1603–1610 (2006)
Gruschke, S. et al. Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. J. Biol. Chem. 285, 19022–19028 (2010)
Jia, L. et al. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22, 6438–6447 (2003)
Agrawal, R. K. & Sharma, M. R. Structural aspects of mitochondrial translational apparatus. Curr. Opin. Struct. Biol. 22, 797–803 (2012)
Pearce, S., Nezich, C. L. & Spinazzola, A. Mitochondrial diseases: translation matters. Mol. Cell. Neurosci. 55, 1–12 (2013)
Sharma, M. R. et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108 (2003)
Mears, J. A. et al. A structural model for the large subunit of the mammalian mitochondrial ribosome. J. Mol. Biol. 358, 193–212 (2006)
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M. & Ban, N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 37, 189–198 (2012)
Walzthoeni, T., Leitner, A., Stengel, F. & Aebersold, R. Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 23, 252–260 (2013)
Polikanov, Y. S., Blaha, G. M. & Steitz, T. A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 (2012)
Smirnov, A., Entelis, N., Martin, R. P. & Tarassov, I. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev. 25, 1289–1305 (2011)
Kelley, L. A. & Sternberg, M. J. E. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371 (2009)
Richter, R. et al. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 29, 1116–1125 (2010)
Handa, Y. et al. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J. Mol. Biol. 404, 260–273 (2010)
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008)
Gagnon, M. G., Seetharaman, S. V., Bulkley, D. & Steitz, T. A. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335, 1370–1372 (2012)
Spirina, O. et al. Heart-specific splice-variant of a human mitochondrial ribosomal protein (mRNA processing; tissue specific splicing). Gene 261, 229–234 (2000)
Carroll, C. J. et al. Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J. Med. Genet. 50, 151–159 (2013)
Liu, M. & Spremulli, L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 275, 29400–29406 (2000)
Schneider, H. C. et al. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371, 768–774 (1994)
Weiss, C. et al. Domain structure and lipid interaction of recombinant yeast Tim44. Proc. Natl Acad. Sci. USA 96, 8890–8894 (1999)
Preuss, M. et al. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 153, 1085–1096 (2001)
Cui, W., Josyula, R., Li, J., Fu, Z. & Sha, B. Membrane binding mechanism of yeast mitochondrial peripheral membrane protein TIM44. Protein Pept. Lett. 18, 718–725 (2011)
Stiburek, L. et al. Knockdown of human Oxa1l impairs the biogenesis of F1Fo-ATP synthase and NADH:ubiquinone oxidoreductase. J. Mol. Biol. 374, 506–516 (2007)
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996)
Loerke, J., Giesebrecht, J. & Spahn, C. M. T. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 483, 161–177 (2010)
Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Gabashvili, I. S. et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100, 537–549 (2000)
Leitner, A. et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012)
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnol. 30, 918–920 (2012)
Rinner, O. et al. Identification of cross-linked peptides from large sequence databases. Nature Methods 5, 315–318 (2008)
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Nakao, A., Yoshihama, M. & Kenmochi, N. RPG: The Ribosomal Protein Gene database. Nucleic Acids Res. 32, D168–D170 (2004)
UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–D219 (2011)
Walzthoeni, T. et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nature Methods 9, 901–903 (2012)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Cannone, J. J. et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2 (2002)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2, 2366–2382 (2007)
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001)
Josyula, R., Jin, Z., Fu, Z. & Sha, B. Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J. Mol. Biol. 359, 798–804 (2006)
Acknowledgements
Cryo-EM data were collected at the electron microscopy facility of ETH Zurich (EMEZ) and at FEI Company Eindhoven. We thank P. Tittmann, F. de Haas and K. Sader for support. We acknowledge the use of computing infrastructure provided by the Central Information Technology Services of ETH Zurich. This work was supported by the Swiss National Science Foundation (SNSF), the National Center of Excellence in Research (NCCR) Structural Biology program of the SNSF, European Research Council (ERC) grant 250071 under the European Community’s Seventh Framework Programme (to N.B.), the Commission of the European Communities through the PROSPECTS consortium (EU FP7 projects 201648, 233226) (R.A.) and the European Research Council (ERC-2008-AdG 233226) (R.A.).
Author information
Authors and Affiliations
Contributions
F.V.-H., J.P.E. and N.B. initiated the project; F.V.-H. and J.P.E. established the purification procedures. F.V.-H., B.J.G. and P.B. performed preparation of the mitoribosomes. B.J.G., P.B. and D.B. prepared cryo-EM samples. D.B. acquired the cryo-EM data. B.J.G., D.B. and P.B. calculated the cryo-EM reconstructions. M.L., B.J.G., P.B., D.B. and N.B. interpreted the structure. A.L. performed CX-MS experiments in the laboratory of R.A. All authors contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
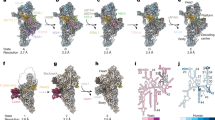
Extended Data Figure 1 Refinement of the 39S mitoribosomal subunit structure.
a, Fourier Shell Correlation (FSC) curve of the cryo-EM reconstruction of the 39S mitoribosomal subunit. The resolution estimate is 4.9 Å according to the FSC = 0.143 criterion (gold standard FSC)38. b, Multiparticle refinement of 39S subunit data sets obtained using the FEI Falcon I and Falcon II direct electron detectors. Particle numbers in the classes are indicated below the volumes. Class 1 was used for further refinement of the structure of the 39S subunit to higher resolution.
Extended Data Figure 2 Location of rRNA and proteins in the 39S subunit.
a, b, View from the solvent side (a) and from the subunit interface side (b) of the 39S mitoribosomal subunit cryo-EM map, segmented into ribosomal proteins (gold) and rRNA (light blue). CP, central protuberance; L1, L1 stalk.
Extended Data Figure 3 Reductive evolution of the mitochondrial 16S rRNA.
Deletions of rRNA secondary structure elements in mitochondrial 16S rRNA are indicated in red on the secondary structure diagram of the Escherichia coli 23S rRNA. Segments for which the precise locations of deletions cannot be deduced from our cryo-EM map (the L1 stalk, a part of domain I rRNA, and the connection to the L7/L12 stalk) are indicated in grey. Depiction is based on the secondary structure of the bacterial 23S rRNA53 (template obtained from the Noller laboratory web page http://rna.ucsc.edu/rnacenter/noller_lab.html).
Extended Data Figure 4 Secondary structure diagram of the S. scrofa large mitoribosomal subunit rRNA.
Domains are coloured according to Fig. 2 and labelled by Roman numerals. Nucleotides not built in our structure are printed in grey. Bacterial secondary structure elements missing in mammalian mitoribosomes are schematically indicated in red. Watson–Crick base pairs are indicated by lines (−), G•U base pairs by dots, and nonstandard base pairs by rings. Depiction is based on the secondary structure of bacterial 23S rRNA53 (template obtained from the Noller lab web page http://rna.ucsc.edu/rnacenter/noller_lab.html).
Extended Data Figure 5 Presence of a second rRNA molecule in the 39S mitoribosomal subunit.
a, Depiction of the mitoribosomal central protuberance RNA (dark blue) and the neighbouring protein MRPL18 (cyan). b, Depiction of the bacterial 5S rRNA (orange) in complex with ribosomal protein L18 (khaki; PDB ID 3V2D). c, Overlay of a and b according to the homologous proteins L18 and MRPL18. d, Overview of the 39S subunit with the fitted RNAs for orientation. e, In bacterial ribosomes, the 5S rRNA connects the central protuberance to the main body of the subunit (the contact region is indicated by an arrow). However, no density for this connection can be seen in the cryo-EM map of the 39S subunit (grey). f, In the mitochondrial 39S subunit, a long α-helical protein density corresponding to MRPL52 (gold) instead of the 5S rRNA connects the mitoribosomal central protuberance (MRPL38, purple) to the subunit body.
Extended Data Figure 6 Quality of the fits of newly positioned 39S mitoribosomal proteins in the cryo-EM density.
Depiction of the cryo-EM density (first column), fits of the unchanged homology models (second column), and fits of the adjusted final models (third column) of MRPL45 (a–c), MRPL44 (d–f), MRPL39 (g–i), MRPL38 (j–l), ICT1 (m–o) and MRPL49 (p–r). The 39S cryo-EM map is shown at two threshold levels (red and grey, respectively).
Extended Data Figure 7 Secondary structure prediction for MRPL52.
The secondary structure prediction (output from Phyre2)19 shows a very long (approximately 50 amino acid) α-helix. Blue stars indicate crosslinks of the modelled domain of MRPL38 to MRPL52, as observed in CX-MS experiments.
Extended Data Figure 8 The polypeptide tunnel exit of the mitoribosomal 39S subunit.
a, b, Comparison of the conserved ring of proteins around the polypeptide tunnel exit (indicated by an asterisk). a, The polypeptide tunnel exit in bacteria17 (PDB ID 3V2D). b, The polypeptide tunnel exit in the porcine mitoribosome. The ring of proteins around the tunnel exit is conserved. c, Additional mitoribosome-specific proteins and protein extensions are also located close to the tunnel exit. The newly identified MRPL39, MRPL44 and MRPL45 are shown in green, red and blue, respectively.
Extended Data Figure 9 The binding sites of the newly docked mitoribosome-specific proteins MRPL44 and MRPL45.
Crosslinks are indicated by double arrows (see also Supplementary Table 3) unless both crosslinking residues have been modelled in the structure. a, MRPL44 (dark red) has been crosslinked to MRPL20 (gold). The crosslinked sites on MRPL44 and MRPL20 are indicated by green and blue spheres, respectively. b, MRPL45 (dark blue) has been crosslinked to MRPL24 (gold) and MRPL23 (orange) in S. scrofa, as well as MRPL22 (pink) and MRPL39 (green) in B. taurus. For all of these crosslinks, at least one crosslink site is located on mitochondria-specific extensions that could not be modelled, and therefore these crosslinks cannot be mapped precisely in the structure. c, Mutation in the human homologue of MRPL44 in mitochondrial infantile cardiomyopathy25. The L156R mutation (green) maps to a residue not involved in 39S binding. The affected residue is located between two α-helices of the RNase domain, and its mutation probably perturbs the structure of MRPL44 and its capacity to bind the 39S subunit.
Extended Data Figure 10 MRPL45 may act as a membrane anchor for the 39S subunit.
a, MRPL45 (blue) may anchor the 39S subunit (grey) to the mitochondrial inner membrane. The membrane surface is tentatively indicated by a horizontal line. b, The structure of the C-terminal domain of TIM44 (ref. 54) (PDB ID 2FXT) with its putative membrane-interacting segment30 is indicated in red. c, The part of MRPL45 corresponding to the putative membrane anchoring segment of TIM44 is shown in red.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3 and additional references. (PDF 207 kb)
Rights and permissions
About this article
Cite this article
Greber, B., Boehringer, D., Leitner, A. et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 (2014). https://doi.org/10.1038/nature12890
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12890
This article is cited by
-
Knockdown of MRPL35 promotes cell apoptosis and inhibits cell proliferation in non-small-cell lung cancer
BMC Pulmonary Medicine (2023)
-
GTPBP8 is required for mitoribosomal biogenesis and mitochondrial translation
Cellular and Molecular Life Sciences (2023)
-
Puf6 primes 60S pre-ribosome nuclear export at low temperature
Nature Communications (2021)
-
Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1
Communications Biology (2021)
-
Mechanisms and regulation of protein synthesis in mitochondria
Nature Reviews Molecular Cell Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.