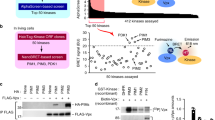
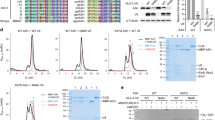
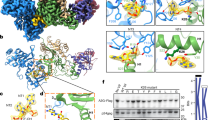
Abstract
Lentiviruses contain accessory genes that have evolved to counteract the effects of host cellular defence proteins that inhibit productive infection. One such restriction factor, SAMHD1, inhibits human immunodeficiency virus (HIV)-1 infection of myeloid-lineage cells1,2 as well as resting CD4+ T cells3,4 by reducing the cellular deoxynucleoside 5′-triphosphate (dNTP) concentration to a level at which the viral reverse transcriptase cannot function5,6. In other lentiviruses, including HIV-2 and related simian immunodeficiency viruses (SIVs), SAMHD1 restriction is overcome by the action of viral accessory protein x (Vpx) or the related viral protein r (Vpr) that target and recruit SAMHD1 for proteasomal degradation7,8. The molecular mechanism by which these viral proteins are able to usurp the host cell’s ubiquitination machinery to destroy the cell’s protection against these viruses has not been defined. Here we present the crystal structure of a ternary complex of Vpx with the human E3 ligase substrate adaptor DCAF1 and the carboxy-terminal region of human SAMHD1. Vpx is made up of a three-helical bundle stabilized by a zinc finger motif, and wraps tightly around the disc-shaped DCAF1 molecule to present a new molecular surface. This adapted surface is then able to recruit SAMHD1 via its C terminus, making it a competent substrate for the E3 ligase to mark for proteasomal degradation. The structure reported here provides a molecular description of how a lentiviral accessory protein is able to subvert the cell’s normal protein degradation pathway to inactivate the cellular viral defence system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011)
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011)
Baldauf, H. M. et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nature Med. 18, 1682–1689 (2012)
Descours, B. et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87 (2012)
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011)
Lahouassa, H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature Immunol. 13, 223–228 (2012)
Lim, E. S. et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 (2012)
Ahn, J. et al. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 (2012)
Morellet, N., Bouaziz, S., Petitjean, P. & Roques, B. P. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 327, 215–227 (2003)
Wei, W. et al. A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell. Microbiol. 14, 1745–1756 (2012)
Srivastava, S. et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4, e1000059 (2008)
Bergamaschi, A. et al. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A–DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83, 4854–4860 (2009)
DeLucia, M., Mehrens, J., Wu, Y. & Ahn, J. HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation. J. Biol. Chem. 288, 19116–19126 (2013)
Bennett, E. J., Bence, N. F., Jayakumar, R. & Kopito, R. R. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell 17, 351–365 (2005)
Scrima, A. et al. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell 135, 1213–1223 (2008)
Fischer, E. S. et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011)
Yan, J. et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 (2013)
Angers, S. et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443, 590–593 (2006)
Li, T., Robert, E. I., van Breugel, P. C., Strubin, M. & Zheng, N. A promiscuous α-helical motif anchors viral hijackers and substrate receptors to the CUL4–DDB1 ubiquitin ligase machinery. Nature Struct. Mol. Biol. 17, 105–111 (2010)
Fregoso, O. I. et al. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 9, e1003496 (2013)
Wen, X., Duus, K. M., Friedrich, T. D. & de Noronha, C. M. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282, 27046–27057 (2007)
Schröfelbauer, B., Hakata, Y. & Landau, N. R. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl Acad. Sci. USA 104, 4130–4135 (2007)
Le Rouzic, E. et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4–DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 (2007)
Hrecka, K. et al. Lentiviral Vpr usurps Cul4–DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl Acad. Sci. USA 104, 11778–11783 (2007)
Barnitz, R. A., Chaigne-Delalande, B., Bolton, D. L. & Lenardo, M. J. Exposed hydrophobic residues in human immunodeficiency virus type 1 Vpr helix-1 are important for cell cycle arrest and cell death. PLoS ONE 6, e24924 (2011)
Doublié, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997)
Zhao, Y., Chapman, D. A. & Jones, I. M. Improving baculovirus recombination. Nucleic Acids Res. 31, e6 (2003)
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012)
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004)
Yap, M. W., Nisole, S., Lynch, C. & Stoye, J. P. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl Acad. Sci. USA 101, 10786–10791 (2004)
Bock, M., Bishop, K. N., Towers, G. & Stoye, J. P. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74, 7422–7430 (2000)
Groom, H. C. et al. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology 7, 10 (2010)
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996)
Bainbridge, J. W. et al. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8, 1665–1668 (2001)
Sundström, C. & Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17, 565–577 (1976)
Gallois-Montbrun, S. et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81, 2165–2178 (2007)
Nègre, D. et al. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7, 1613–1623 (2000)
Acknowledgements
We thank L. Haire and R. Ogrodowicz for help with crystallization, I. Jones for the provision of the modified A. californica baculovirus bacmid, BAC10:1629KO, S. Smerdon and S. Gamblin for comments on the manuscript. We gratefully acknowledge the Diamond Light Source for synchrotron access (grant no. 7707). This work was supported by the UK Medical Research Council, file references U117565647 (I.A.T.), U117592729 (K.N.B.) and U117512710 (J.P.S.); the Wellcome Trust, ref. 084955 (K.N.B.); and by an EMBO long-term fellowship co-funded by the European Commission Marie Curie Actions (EMBOCOFUND2010, GA-2010-267146) (D.S.).
Author information
Authors and Affiliations
Contributions
D.S., H.C.T.G., V.C.B., E.C. and P.A.W. performed experiments. D.S., H.C.T.G., V.C.B., E.C., P.A.W., J.P.S., K.N.B. and I.A.T. contributed to experimental design, data analysis and manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Experimental electron density.
Experimental electron density observed after solvent flattening for DCAF1, Vpxsm and SAMHD1 is shown as light blue wireframe, contoured at 1σ. The backbone Cα traces of the final refined protein structure are shown in green ribbon representation.
Extended Data Figure 2 Model of the hijacked CUL4A–DDB1–ROC1 E3 ubiquitin ligase complex.
Vpxsm (blue) bound to the substrate-specificity module DCAF1-CtD (grey) is shown in cartoon representation as in Fig. 1c. SAMHD1-CtD is shown as red spheres. The DDB1 adaptor is shown as green cartoon. The inset to the right shows the superposition of the DDB2 helical hairpin (H-box, orange), which inserts into the binding groove created by DDB1 β-propellers 1 and 3 (BPA, BPC), and the N terminus of DCAF1-CtD presented in this study. The CUL4A scaffold is represented as an orange semi-transparent surface, the ROC1 RING module as purple spheres. Owing to the conformational freedom of the DDB1–CUL4A connection, the two most extreme conformations of CUL4A with respect to DDB1 available in the PDB were modelled. See Methods for modelling procedures and PDB accessions. The model clearly shows that in both extreme CUL4 conformations, the ROC1 RING finger (purple spheres) is well positioned to reach the SAMHD1 protein, which would be attached at the N-terminal end of the SAMHD1-CtD. The SAMHD1 globular fold is probably mobile with respect to the fixed position of SAMHD1-CtD owing to the flexibility of the sequence stretch between SAMHD1 residues 583 (the last ordered residue of PDB accession 3U1N5) and 606 (the first ordered residue of SAMHD1-CtD presented here).
Extended Data Figure 3 Analogous mechanism of restriction factor counteraction in SIV/HIV-2 and HIV-1 Vpx and Vpr.
SAMHD1 provides a potent post-entry block against immunodeficiency viruses in non-cycling cells. Its dNTP-triphosphohydrolase activity lowers the cellular dNTP pool, preventing viral reverse transcription. HIV-2/SIVs use their Vpx and Vpr accessory proteins to modify the host cell’s CUL4A–DDB1–DCAF1 ubiquitin ligase specificity towards SAMHD1, resulting in its proteasomal degradation and ultimately raising dNTP levels, making the cells permissive to viral replication. Sequence similarity and comparative functional analysis suggest that the ancestral HIV-1 accessory protein Vpr uses a similar mechanism to exploit the CUL4A–DDB1–DCAF1 system to induce proteasomal degradation of an as yet undiscovered cellular factor whose absence causes cell cycle arrest in the G2 phase, promoting viral replication and pathogenesis in vivo.
Rights and permissions
About this article
Cite this article
Schwefel, D., Groom, H., Boucherit, V. et al. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505, 234–238 (2014). https://doi.org/10.1038/nature12815
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12815
This article is cited by
-
Centrosome amplification and aneuploidy driven by the HIV-1-induced Vpr•VprBP•Plk4 complex in CD4+ T cells
Nature Communications (2024)
-
DCAF1-based PROTACs with activity against clinically validated targets overcoming intrinsic- and acquired-degrader resistance
Nature Communications (2024)
-
Attenuation of reverse transcriptase facilitates SAMHD1 restriction of HIV-1 in cycling cells
Retrovirology (2023)
-
HIV-2/SIV Vpx antagonises NF-κB activation by targeting p65
Retrovirology (2022)
-
HIV-1 Vpr activates host CRL4-DCAF1 E3 ligase to degrade histone deacetylase SIRT7
Virology Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.