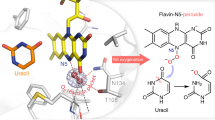
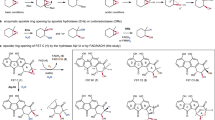
Abstract
Flavoproteins catalyse a diversity of fundamental redox reactions and are one of the most studied enzyme families1,2. As monooxygenases, they are universally thought to control oxygenation by means of a peroxyflavin species that transfers a single atom of molecular oxygen to an organic substrate1,3,4. Here we report that the bacterial flavoenzyme EncM5,6 catalyses the peroxyflavin-independent oxygenation–dehydrogenation dual oxidation of a highly reactive poly(β-carbonyl). The crystal structure of EncM with bound substrate mimics and isotope labelling studies reveal previously unknown flavin redox biochemistry. We show that EncM maintains an unexpected stable flavin-oxygenating species, proposed to be a flavin-N5-oxide, to promote substrate oxidation and trigger a rare Favorskii-type rearrangement that is central to the biosynthesis of the antibiotic enterocin. This work provides new insight into the fine-tuning of the flavin cofactor in offsetting the innate reactivity of a polyketide substrate to direct its efficient electrocyclization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Accessions
GenBank/EMBL/DDBJ
Protein Data Bank
Data deposits
The EncM sequence is deposited in GenBank under accession number AAF81732.1. The structures for proteins described in this paper have been deposited in the Protein Data Bank under accession numbers 3W8W (apo-EncM), 3W8X (EncM with bound 26) and 3W8Z (EncM with bound 4). Data for the crystallized substrate analogues are deposited in the Cambridge Crystallographic Data Centre under accession numbers CCDC 922822 (4), CCDC 922821 (10) and CCDC 949270 (26).
References
Walsh, C. T. & Wencewicz, T. A. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 30, 175–200 (2012)
Chaiyen, P., Fraaije, M. W. & Mattevi, A. The enigmatic reaction of flavins with oxygen. Trends Biochem. Sci. 37, 373–380 (2012)
Massey, V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 269, 22459–22462 (1994)
Palfey, B. A. & McDonald, C. A. Control of catalysis in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 493, 26–36 (2010)
Xiang, L., Kalaitzis, J. A. & Moore, B. S. EncM, a versatile enterocin biosynthetic enzyme involved in Favorskii oxidative rearrangement, aldol condensation, and heterocycle-forming reactions. Proc. Natl Acad. Sci. USA 101, 15609–15614 (2004)
Cheng, Q., Xiang, L., Izumikawa, M., Meluzzi, D. & Moore, B. S. Enzymatic total synthesis of enterocin polyketides. Nature Chem. Biol. 3, 557–558 (2007)
Piel, J. et al. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate ‘Streptomyces maritimus’: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 7, 943–955 (2000)
Seto, H., Sato, T., Urano, S., Uzawa, J. & Yonehara, H. Utilization of 13C–13C coupling in structural and biosynthetic studies. VII. The structure and biosynthesis of vulgamycin. Tetrahedr. Lett. 48, 4367–4370 (1976)
Hertweck, C. et al. Context-dependent behavior of the enterocin iterative polyketide synthase; a new model for ketoreduction. Chem. Biol. 11, 461–468 (2004)
Hertweck, C., Luzhetskyy, A., Rebets, Y. & Bechthold, A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190 (2007)
Koetter, J. W. & Schulz, G. E. Crystal structure of 6-hydroxy-d-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 352, 418–428 (2005)
Huang, C. H. et al. Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8α-N1-histidyl FAD. J. Biol. Chem. 280, 38831–38838 (2005)
Alexeev, I., Sultana, A., Mantsala, P., Niemi, J. & Schneider, G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl Acad. Sci. USA 104, 6170–6175 (2007)
Crosby, J. & Crump, M. P. The structural role of the carrier protein–active controller or passive carrier. Nat. Prod. Rep. 29, 1111–1137 (2012)
Baumann, K., Bacher, M., Damont, A. & Steck, A. Selective transformation of ascomycin into 11-epi-ascomycin. Tetrahedr. Lett. 45, 549–551 (2004)
Takikawa, H., Takada, A., Hikita, K. & Suzuki, K. Formation of α-hydroxy-β-diketones through hydroxylation of isoxazolium salts: stereoselective approach to angular cis-diols in polycyclic systems. Angew. Chem. Int. Edn Engl. 47, 7446–7449 (2008)
Entsch, B., Ballou, D. P. & Massey, V. Flavin–oxygen derivatives involved in hydroxylation by p-hydroxybenzoate hydroxylase. J. Biol. Chem. 251, 2550–2563 (1976)
Entsch, B. & Ballou, D. P. Purification, properties, and oxygen reactivity of p-hydroxybenzoate hydroxylase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 999, 313–322 (1989)
Thotsaporn, K., Chenprakhon, P., Sucharitakul, J., Mattevi, A. & Chaiyen, P. Stabilization of C4a-hydroperoxyflavin in a two-component flavin-dependent monooxygenase is achieved through interactions at flavin N5 and C4a atoms. J. Biol. Chem. 286, 28170–28180 (2011)
Valton, J., Mathevon, C., Fontecave, M., Niviere, V. & Ballou, D. P. Mechanism and regulation of the two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. J. Biol. Chem. 283, 10287–10296 (2008)
Rastetter, W. H., Gadek, T. R., Tane, J. P. & Frost, J. W. Oxidations and oxygen transfers effected by a flavin N(5)-oxide. A model for flavin-dependent monooxygenases. J. Am. Chem. Soc. 101, 2228–2231 (1979)
Orf, H. W. & Dolphin, D. Oxaziridines as possible intermediates in flavin monooxygenases. Proc. Natl Acad. Sci. USA 71, 2646–2650 (1974)
Walsh, C. et al. Chemical and enzymatic properties of riboflavin analogues. Biochemistry 17, 1942–1951 (1978)
Garman, E. F. & Owen, R. L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 62, 32–47 (2006)
van Berkel, W. J., Kamerbeek, N. M. & Fraaije, M. W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689 (2006)
Izumikawa, M., Cheng, Q. & Moore, B. S. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J. Am. Chem. Soc. 128, 1428–1429 (2006)
Kalaitzis, J. A., Izumikawa, M., Xiang, L., Hertweck, C. & Moore, B. S. Mutasynthesis of enterocin and wailupemycin analogues. J. Am. Chem. Soc. 125, 9290–9291 (2003)
Han, L., Lobo, S. & Reynolds, K. A. Characterization of β-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J. Bacteriol. 180, 4481–4486 (1998)
Jez, J. M., Ferrer, J. L., Bowman, M. E., Dixon, R. A. & Noel, J. P. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39, 890–902 (2000)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newsl. Prot. Crystallogr. 26, 27–33 (1992)
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010)
Morris, R. J., Perrakis, A. & Lamzin, V. S. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 58, 968–975 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific LLC, 2002)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006)
Tovchigrechko, A. & Vakser, I. A. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 34, W310–W314 (2006)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Acknowledgements
We thank M. Bowman for technical assistance; Y. Su for mass spectrometric measurements; D.-H. Huang and L. Pasternack for NMR spectroscopic assistance; A. Rheingold for X-ray crystallographic analysis; and C. Hertweck for establishing the synthesis of 26. This research was supported by US National Institutes of Health grant R01AI47818 to B.S.M., National Science Foundation (NSF) awards EEC-0813570 and MCB-0645794 and the Howard Hughes Medical Institute for grants to J.P.N., NSF grant CHE-1213620 to B.P., and fellowships to R.T. from the Deutsche Forschungsgemeinschaft (TE 931/1-1) and to A.M. from the Japan Society for the Promotion of Science (21-644).
Author information
Authors and Affiliations
Contributions
R.T., A.M., Q.M., F.S., G.L. and J.P.N. performed research. All authors designed research and analysed data. R.T. and B.M. wrote the paper. R.T., A.M., Q.M. and F.S. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Information, which is divided into parts 1 and 2: (1) Supplementary Figures 1-26, Supplementary Tables 1-17, Supplementary Text and Data (pages 3-58) and (2) NMR spectra of all chemically synthesized compounds (pages 60-86), see Contents 1 and 2 (page 59) for more details. (PDF 10595 kb)
Rights and permissions
About this article
Cite this article
Teufel, R., Miyanaga, A., Michaudel, Q. et al. Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 503, 552–556 (2013). https://doi.org/10.1038/nature12643
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12643
This article is cited by
-
Triepoxide formation by a flavin-dependent monooxygenase in monensin biosynthesis
Nature Communications (2023)
-
A flavin-monooxygenase catalyzing oxepinone formation and the complete biosynthesis of vibralactone
Nature Communications (2023)
-
Enzymatic spiroketal formation via oxidative rearrangement of pentangular polyketides
Nature Communications (2021)
-
A host–guest approach to combining enzymatic and artificial catalysis for catalyzing biomimetic monooxygenation
Nature Communications (2020)
-
Flavin doesn’t put all oxygens in one basket
Nature Chemical Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.