Abstract
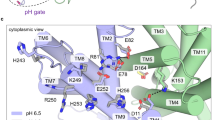
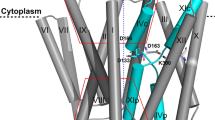
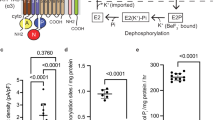
Sodium/proton (Na+/H+) antiporters, located at the plasma membrane in every cell, are vital for cell homeostasis1. In humans, their dysfunction has been linked to diseases, such as hypertension, heart failure and epilepsy, and they are well-established drug targets2. The best understood model system for Na+/H+ antiport is NhaA from Escherichia coli1,3, for which both electron microscopy and crystal structures are available4,5,6. NhaA is made up of two distinct domains: a core domain and a dimerization domain. In the NhaA crystal structure a cavity is located between the two domains, providing access to the ion-binding site from the inward-facing surface of the protein1,4. Like many Na+/H+ antiporters, the activity of NhaA is regulated by pH, only becoming active above pH 6.5, at which point a conformational change is thought to occur7. The only reported NhaA crystal structure so far is of the low pH inactivated form4. Here we describe the active-state structure of a Na+/H+ antiporter, NapA from Thermus thermophilus, at 3 Å resolution, solved from crystals grown at pH 7.8. In the NapA structure, the core and dimerization domains are in different positions to those seen in NhaA, and a negatively charged cavity has now opened to the outside. The extracellular cavity allows access to a strictly conserved aspartate residue thought to coordinate ion binding1,8,9 directly, a role supported here by molecular dynamics simulations. To alternate access to this ion-binding site, however, requires a surprisingly large rotation of the core domain, some 20° against the dimerization interface. We conclude that despite their fast transport rates of up to 1,500 ions per second3, Na+/H+ antiporters operate by a two-domain rocking bundle model, revealing themes relevant to secondary-active transporters in general.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Padan, E. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem. Sci. 33, 435–443 (2008)
Brett, C. L., Donowitz, M. & Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239 (2005)
Taglicht, D., Padan, E. & Schuldiner, S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 266, 11289–11294 (1991)
Hunte, C. et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005)
Williams, K. A., Geldmacher-Kaufer, U., Padan, E., Schuldiner, S. & Kuhlbrandt, W. Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0 Å resolution. EMBO J. 18, 3558–3563 (1999)
Williams, K. A. Three-dimensional structure of the ion-coupled transport protein NhaA. Nature 403, 112–115 (2000)
Kozachkov, L. & Padan, E. Conformational changes in NhaA Na+/H+ antiporter. Mol. Membr. Biol. 30, 90–100 (2012)
Arkin, I. T. et al. Mechanism of Na+/H+ antiporting. Science 317, 799–803 (2007)
Maes, M., Rimon, A., Kozachkov-Magrisso, L., Friedler, A. & Padan, E. Revealing the ligand binding site of NhAa Na+/H+ antiporter and its pH dependence. J. Biol. Chem. 287, 38150–38157 (2012)
West, I. C. & Mitchell, P. Proton/sodium ion antiport in Escherichia coli. Biochem. J. 144, 87–90 (1974)
Taglicht, D., Padan, E. & Schuldiner, S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J. Biol. Chem. 268, 5382–5387 (1993)
Pinner, E., Padan, E. & Schuldiner, S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J. Biol. Chem. 269, 26274–26279 (1994)
Drew, D., Lerch, M., Kunji, E., Slotboom, D. J. & de Gier, J. W. Optimization of membrane protein overexpression and purification using GFP fusions. Nature Methods 3, 303–313 (2006)
Sonoda, Y. et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure 19, 17–25 (2011)
Furrer, E. M., Ronchetti, M. F., Verrey, F. & Pos, K. M. Functional characterization of a NapA Na+/H+ antiporter from Thermus thermophilus. FEBS Lett. 581, 572–578 (2007)
Padan, E. et al. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim. Biophys. Acta 1658, 2–13 (2004)
Rimon, A., Tzubery, T. & Padan, E. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+. J. Biol. Chem. 282, 26810–26821 (2007)
Hisamitsu, T., Ben Ammar, Y., Nakamura, T. Y. & Wakabayashi, S. Dimerization is crucial for the function of the Na+/H+ exchanger NHE1. Biochemistry 45, 13346–13355 (2006)
Appel, M., Hizlan, D., Vinothkumar, K. R., Ziegler, C. & Kuhlbrandt, W. Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J. Mol. Biol. 388, 659–672 (2009)
Goswami, P. et al. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 30, 439–449 (2011)
Kuwabara, N., Inoue, H., Tsuboi, Y., Nakamura, N. & Kanazawa, H. The fourth transmembrane domain of the Helicobacter pylori Na+/H+ antiporter NhaA faces a water-filled channel required for ion transport. J. Biol. Chem. 279, 40567–40575 (2004)
Mager, T., Rimon, A., Padan, E. & Fendler, K. Transport Mechanism and pH Regulation of the Na+/H+ Antiporter NhaA from Escherichia coli. J. Biol. Chem. 286, 23570–23581 (2011)
Screpanti, E. & Hunte, C. Discontinuous membrane helices in transport proteins and their correlation with function. J. Struct. Biol. 159, 261–267 (2007)
Hu, N. J., Iwata, S., Cameron, A. D. & Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408–411 (2011)
Noumi, T., Inoue, H., Sakurai, T., Tsuchiya, T. & Kanazawa, H. Identification and characterization of functional residues in a Na+/H+ antiporter (NhaA) from Escherichia coli by random mutagenesis. J. Biochem. 121, 661–670 (1997)
Vinothkumar, K. R., Smits, S. H. & Kuhlbrandt, W. pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 24, 2720–2729 (2005)
Reyes, N., Ginter, C. & Boudker, O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462, 880–885 (2009)
Schushan, M. et al. A model-structure of a periplasm-facing state of the NhaA antiporter suggests the molecular underpinnings of pH-induced conformational changes. J. Biol. Chem. 287, 18249–18261 (2012)
Drew, D. E., von Heijne, G., Nordlund, P. & de Gier, J. W. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 507, 220–224 (2001)
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006)
Wagner, S. et al. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl Acad. Sci. USA 105, 14371–14376 (2008)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Drew, D. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nature Protocols 3, 784–798 (2008)
Slotboom, D. J., Duurkens, R. H., Olieman, K. & Erkens, G. B. Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 (2008)
Wiedenmann, A., Dimroth, P. & von Ballmoos, C. Functional asymmetry of the F0 motor in bacterial ATP synthases. Mol. Microbiol. 72, 479–490 (2009)
Ishmukhametov, R. R., Galkin, M. A. & Vik, S. B. Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1Fo ATP synthase. Biochim. Biophys. Acta 1706, 110–116 (2005)
Wiedenmann, A., Dimroth, P. & von Ballmoos, C. Δψ and ΔpH are equivalent driving forces for proton transport through isolated F0 complexes of ATP synthases. Biochim. Biophys. Acta 1777, 1301–1310 (2008)
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Knight, S. D. RSPS version 4.0: a semi-interactive vector-search program for solving heavy-atom derivatives. Acta Crystallogr. D 56, 42–47 (2000)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Cowtan, K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Prot. Crystallogr. 31, 34–38 (1994)
Jones, T. A., Kjeldgaard, M., Charles, W. C., Jr & Robert, M. S. Electron-density map interpretation. Methods Enzymol. 277, 173–208 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Kleywegt, G. J. & Jones, T. A. A super position. Joint CCP4 ESF-EACBM Newslett. Prot. Crystallogr. 31, 9–14 (1994)
Delano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002)
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D 60, 2288–2294 (2004)
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008)
MacKerell, A. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998)
Mackerell, J. A. D., Jr, Feig, M. & Brooks, C. L., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 (2004)
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Stansfeld, P. J. & Sansom, M. S. P. From coarse grained to atomistic: a serial multiscale approach to membrane protein simulations. J. Chem. Theory Comput. 7, 1157–1166 (2011)
Bond, P. J., Wee, C. L. & Sansom, M. S. P. Coarse-grained molecular dynamics simulations of the energetics of helix insertion into a lipid bilayer. Biochemistry 47, 11321–11331 (2008)
Scott, K. A. et al. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure 16, 621–630 (2008)
Li, H., Robertson, A. D. & Jensen, J. H. Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 (2005)
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007)
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981)
Hess, B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008)
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8592 (1995)
Miyamoto, S. & Kollman, P. A. SETTLE: An analytical version of the SHAKE and RATTLE algorithms for rigid water models. J. Comput. Chem. 13, 952–962 (1992)
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011)
Humphrey, W., Dalke, A. & Schulten, K. VMD—visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Dahl, A. C. E., Chavent, M. & Sansom, M. S. P. Bendix: Intuitive helix geometry analysis and abstraction for VMD. Bioinformatics 28, 2193–2194 (2012)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Acknowledgements
We are grateful to D. Slotboom for critical reading of the manuscript and N.-J. Hu for assistance in data collection. Data were collected at the European Synchrotron Radiation Facility and Diamond Light Source, with excellent assistance from beamline scientists. This work was funded by grants from the Medical Research Council (MRC grant G0900990 to A.D.C. and D.D.), the Swedish Research Council (to C.v.B. and D.D.) and the BBSRC (BB/G02325/1 to S.I.). The authors are grateful for the use of the Membrane Protein Laboratory funded by the Wellcome Trust (grant 062164/Z/00/Z) at the Diamond Light Source Limited and The Centre for Biomembrane Research (CBR), supported by the Swedish Foundation for Strategic Research. Computer simulations were partially run on XSEDE resources (grant TG-MCB120151 to O.B.). C.L. was a recipient of a BBSRC-funded PhD scholarship, H.J.K. a Human Frontiers Science Program (HFSP) postdoctoral fellowship, and D.D. acknowledges the support from The Royal Society through the University Research Fellow (URF) scheme.
Author information
Authors and Affiliations
Contributions
A.D.C. and D.D. designed the project. Cloning, expression screening, protein purification and crystallization were carried out by C.L. and D.D. with assistance from H.J.K., S.N., S.I. and A.D.C. Data collection and structural determination were carried out by C.L., D.D. and A.D.C. Experiments for functional analysis were designed by C.v.B. and D.D. and carried out by C.v.B., C.L., P.U. and D.D. Molecular dynamics simulations were carried out by D.L.D. and O.B. A.D.C. and D.D. wrote the manuscript with contributions from C.L., H.J.K., C.v.B. and O.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8, Supplementary Tables 1-2 and additional references. (PDF 9005 kb)
Alternating access model of Na+/H+ antiport.
Video morphing between the outward-facing NapA crystal structure and a model created by superposing the NapA Core and Dimerisation domains separately onto the NhaA structure. The residues mentioned in the text are shown in a stick representation. Colouring as in Figure 3. (MOV 5509 kb)
Alternating access model of Na+/H+ antiport.
Video morphing between the outward-facing NapA crystal structure and a model created by superposing the NapA Core and Dimerisation domains separately onto the NhaA structure. The residues mentioned in the text are shown in a stick representation. Colouring as in Figure 3. (MOV 8207 kb)
Rights and permissions
About this article
Cite this article
Lee, C., Kang, H., von Ballmoos, C. et al. A two-domain elevator mechanism for sodium/proton antiport. Nature 501, 573–577 (2013). https://doi.org/10.1038/nature12484
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12484
This article is cited by
-
Ion and lipid orchestration of secondary active transport
Nature (2024)
-
Architecture and autoinhibitory mechanism of the plasma membrane Na+/H+ antiporter SOS1 in Arabidopsis
Nature Communications (2023)
-
Determining small-molecule permeation through lipid membranes
Nature Protocols (2022)
-
Crystal structure of the Na+/H+ antiporter NhaA at active pH reveals the mechanistic basis for pH sensing
Nature Communications (2022)
-
Structural insights into the HBV receptor and bile acid transporter NTCP
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.