Abstract
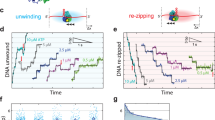
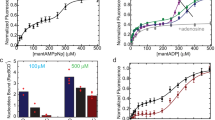
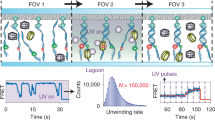
Single-molecule studies can overcome the complications of asynchrony and ensemble-averaging in bulk-phase measurements, provide mechanistic insights into molecular activities, and reveal interesting variations between individual molecules1,2,3. The application of these techniques to the RecBCD helicase of Escherichia coli has resolved some long-standing discrepancies, and has provided otherwise unattainable mechanistic insights into its enzymatic behaviour4,5,6. Enigmatically, the DNA unwinding rates of individual enzyme molecules are seen to vary considerably6,7,8, but the origin of this heterogeneity remains unknown. Here we investigate the physical basis for this behaviour. Although any individual RecBCD molecule unwound DNA at a constant rate for an average of approximately 30,000 steps, we discover that transiently halting a single enzyme–DNA complex by depleting Mg2+-ATP could change the subsequent rates of DNA unwinding by that enzyme after reintroduction to ligand. The proportion of molecules that changed rate increased exponentially with the duration of the interruption, with a half-life of approximately 1 second, suggesting that a conformational change occurred during the time that the molecule was arrested. The velocity after pausing an individual molecule was any velocity found in the starting distribution of the ensemble. We suggest that substrate binding stabilizes the enzyme in one of many equilibrium conformational sub-states that determine the rate-limiting translocation behaviour of each RecBCD molecule. Each stabilized sub-state can persist for the duration (approximately 1 minute) of processive unwinding of a DNA molecule, comprising tens of thousands of catalytic steps, each of which is much faster than the time needed for the conformational change required to alter kinetic behaviour. This ligand-dependent stabilization of rate-defining conformational sub-states results in seemingly static molecule-to-molecule variation in RecBCD helicase activity, but in fact reflects one microstate from the equilibrium ensemble that a single molecule manifests during an individual processive translocation event.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Moffitt, J. R., Chemla, Y. R., Smith, S. B. & Bustamante, C. Recent advances in optical tweezers. Annu. Rev. Biochem. 77, 205–228 (2008)
Ha, T. Single-molecule fluorescence resonance energy transfer. Methods 25, 78–86 (2001)
Xie, X. S. & Lu, H. P. Single-molecule enzymology. J. Biol. Chem. 274, 15967–15970 (1999)
Spies, M. et al. A molecular throttle: the recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell 114, 647–654 (2003)
Spies, M., Amitani, I., Baskin, R. J. & Kowalczykowski, S. C. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131, 694–705 (2007)
Handa, N., Bianco, P. R., Baskin, R. J. & Kowalczykowski, S. C. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after χ recognition. Mol. Cell 17, 745–750 (2005)
Bianco, P. R. et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001)
Dillingham, M. S. & Kowalczykowski, S. C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 (2008)
Lam, S. T., Stahl, M. M., McMilin, K. D. & Stahl, F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77, 425–433 (1974)
Dillingham, M. S., Spies, M. & Kowalczykowski, S. C. RecBCD enzyme is a bipolar DNA helicase. Nature 423, 893–897 (2003)
Taylor, A. F. & Smith, G. R. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423, 889–893 (2003)
Handa, N. et al. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, χ, by RecBCD enzyme. Proc. Natl Acad. Sci. USA 109, 8901–8906 (2012)
Singleton, M. R., Dillingham, M. S., Gaudier, M., Kowalczykowski, S. C. & Wigley, D. B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 (2004)
Dillingham, M. S., Webb, M. R. & Kowalczykowski, S. C. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 280, 37069–37077 (2005)
Frauenfelder, H., McMahon, B. H., Austin, R. H., Chu, K. & Groves, J. T. The role of structure, energy landscape, dynamics, and allostery in the enzymatic function of myoglobin. Proc. Natl Acad. Sci. USA 98, 2370–2374 (2001)
Lu, H. P., Xun, L. & Xie, X. S. Single-molecule enzymatic dynamics. Science 282, 1877–1882 (1998)
Xue, Q. & Yeung, E. S. Differences in the chemical reactivity of individual molecules of an enzyme. Nature 373, 681–683 (1995)
Craig, D. B., Arriaga, E. A., Wong, J. C. Y., Lu, H. & Dovichi, N. J. Studies on single alkaline phosphatase molecules: reaction rate and activation energy of a reaction catalyzed by a single molecule and the effect of thermal denaturation – the death of an enzyme. J. Am. Chem. Soc. 118, 5245–5253 (1996)
Shi, J. et al. Multiple states of the Tyr318Leu mutant of dihydroorotate dehydrogenase revealed by single-molecule kinetics. J. Am. Chem. Soc. 126, 6914–6922 (2004)
Wolynes, P. G., Onuchic, J. N. & Thirumalai, D. Navigating the folding routes. Science 267, 1619–1620 (1995)
Onuchic, J. N., Wolynes, P. G., Luthey-Schulten, Z. & Socci, N. D. Toward an outline of the topography of a realistic protein-folding funnel. Proc. Natl Acad. Sci. USA 92, 3626–3630 (1995)
Dill, K. A., Ozkan, S. B., Shell, M. S. & Weikl, T. R. The protein folding problem. Annu. Rev. Biophys. 37, 289–316 (2008)
Nguyen, H. D. & Hall, C. K. Effect of rate of chemical or thermal renaturation on refolding and aggregation of a simple lattice protein. Biotechnol. Bioeng. 80, 823–834 (2002)
Ikai, A. & Tanford, C. Kinetic evidence for incorrectly folded intermediate states in the refolding of denatured proteins. Nature 230, 100–102 (1971)
Sela, M., White, F. H., Jr & Anfinsen, C. B. Reductive cleavage of disulfide bridges in ribonuclease. Science 125, 691–692 (1957)
Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991)
Ma, B. & Nussinov, R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 14, 652–659 (2010)
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nature Chem. Biol. 5, 789–796 (2009)
Perkins, T. T., Li, H. W., Dalal, R. V., Gelles, J. & Block, S. M. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 86, 1640–1648 (2004)
Amitani, I., Liu, B., Dombrowski, C. C., Baskin, R. J. & Kowalczykowski, S. C. Watching individual proteins acting on single molecules of DNA. Methods Enzymol. 472, 261–291 (2010)
Roman, L. J. & Kowalczykowski, S. C. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 28, 2863–2873 (1989)
Bianco, P. R. & Kowalczykowski, S. C. The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc. Natl Acad. Sci. USA 94, 6706–6711 (1997)
Spies, M., Dillingham, M. S. & Kowalczykowski, S. C. Translocation by the RecB motor is an absolute requirement for χ−recognition and RecA protein loading by RecBCD enzyme. J. Biol. Chem. 280, 37078–37087 (2005)
Kreuzer, K. N. & Jongeneel, C. V. Escherichia coli phage T4 topoisomerase. Methods Enzymol. 100, 144–160 (1983)
Neuman, K. C. & Block, S. M. Optical trapping. Rev. Sci. Instrum. 75, 2787–2809 (2004)
Acknowledgements
We are grateful to members of the laboratory for their comments on this work. S.C.K. was supported by the National Institutes of Health (GM-62653 and GM-64745).
Author information
Authors and Affiliations
Contributions
B.L., R.J.B. and S.C.K. conceived the general ideas, designed the experiments and interpreted the data. B.L. performed experiments. B.L. and S.C.K. analysed the data and wrote the manuscript. R.J.B. passed away on July 3, 2010; this work is dedicated to his collegiality and contributions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-10. (PDF 793 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Liu, B., Baskin, R. & Kowalczykowski, S. DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature 500, 482–485 (2013). https://doi.org/10.1038/nature12333
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12333
This article is cited by
-
Auxiliary ATP binding sites support DNA unwinding by RecBCD
Nature Communications (2022)
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
Rotation tracking of genome-processing enzymes using DNA origami rotors
Nature (2019)
-
Modeling DNA Unwinding by AddAB Helicase–Nuclease and Modulation by Chi Sequences: Comparison with AdnAB and RecBCD
Cellular and Molecular Bioengineering (2019)
-
Dynamic coordination of two-metal-ions orchestrates λ-exonuclease catalysis
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.