Abstract
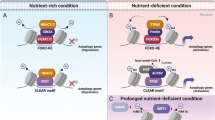
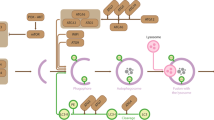
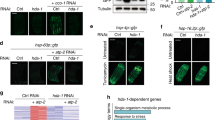
Autophagy is an evolutionarily conserved catabolic process involved in several physiological and pathological processes1,2. Although primarily cytoprotective, autophagy can also contribute to cell death; it is thus important to understand what distinguishes the life or death decision in autophagic cells3. Here we report that induction of autophagy is coupled to reduction of histone H4 lysine 16 acetylation (H4K16ac) through downregulation of the histone acetyltransferase hMOF (also called KAT8 or MYST1), and demonstrate that this histone modification regulates the outcome of autophagy. At a genome-wide level, we find that H4K16 deacetylation is associated predominantly with the downregulation of autophagy-related genes. Antagonizing H4K16ac downregulation upon autophagy induction results in the promotion of cell death. Our findings establish that alteration in a specific histone post-translational modification during autophagy affects the transcriptional regulation of autophagy-related genes and initiates a regulatory feedback loop, which serves as a key determinant of survival versus death responses upon autophagy induction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
21 August 2013
An addition was made to the Acknowledgements section.
References
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008)
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008)
Levine, B. & Yuan, J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679–2688 (2005)
Yang, Z. & Klionsky, D. J. Eaten alive: a history of macroautophagy. Nature Cell Biol. 12, 814–822 (2010)
Chen, Y. & Klionsky, D. J. The regulation of autophagy – unanswered questions. J. Cell Sci. 124, 161–170 (2011)
Lee, I. H. & Finkel, T. Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 284, 6322–6328 (2009)
Lee, I. H. et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl Acad. Sci. USA 105, 3374–3379 (2008)
Yi, C. et al. Function and molecular mechanism of acetylation in autophagy regulation. Science 336, 474–477 (2012)
Lin, S. Y. et al. GSK3–TIP60–ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481 (2012)
Morselli, E. et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615–629 (2011)
Morselli, E. et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 1, e10 (2010)
Vaquero, A., Sternglanz, R. & Reinberg, D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 26, 5505–5520 (2007)
Hajji, N. et al. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene 29, 2192–2204 (2010)
Taipale, M. et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25, 6798–6810 (2005)
Smith, E. R. et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25, 9175–9188 (2005)
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006)
Kind, J. et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133, 813–828 (2008)
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008)
Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011)
Ruthenburg, A. J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011)
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009)
Katoh, H. et al. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol. Cell 44, 770–784 (2011)
Zhou, Y. & Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 15, 1434–1438 (2005)
Fullgrabe, J., Hajji, N. & Joseph, B. Cracking the death code: apoptosis-related histone modifications. Cell Death Differ. 17, 1238–1243 (2010)
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007)
Shechter, D., Dormann, H. L., Allis, C. D. & Hake, S. B. Extraction, purification and analysis of histones. Nature Protocols 2, 1445–1457 (2007)
Burguillos, M. A. et al. Caspase signalling controls microglia activation and neurotoxicity. Nature 472, 319–324 (2011)
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012)
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005)
Cao, Y., Cheong, H., Song, H. & Klionsky, D. J. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J. Cell Biol. 182, 703–713 (2008)
Shintani, T. & Klionsky, D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894 (2004)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Ji, H. et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnol. 26, 1293–1300 (2008)
Saldanha, A. J. Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. &, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
Acknowledgements
We thank G. Mc Inerney, M. Malewicz, S. Orrenius and T. Panaretakis for discussion, and L. Guarente, V. Kaminskyy, M. Komatsu, G. Mc Inerney, M. Panas, R. G. Roeder and L. Xiaoling for reagents and cell lines. J.F. is supported by a fellowship from the Karolinska Institutet Foundations, M.A.L.-D. is partly supported by a Rackham Predoctoral Fellowship and W.L. is supported by breast cancer research Postdoctoral Fellowship Award (BC110381) from the US Department of Defense. This work was supported by a National Institutes of Health grant GM53396 (to D.J.K.) and National Institutes of Health/National Cancer Institute and Department of Defense grants (to M.G.R.), the Swedish Cancer Society, the Swedish Childhood Cancer Foundation (to B.J. and O.H.) and the Swedish Research Council (to B.J.). M.G.R. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
J.F., R.B.S. and B.J. performed experiments in mammalian cells. M.A.L.-D. performed yeast experiments. N.H. performed ChIP-seq. W.L. performed GRO-seq. N.H. and Q.M. performed bioinformatical analysis. J.F., O.H., M.G.R., D.J.K. and B.J. designed the study, and analysed and interpreted the data. The first draft of the paper was written by J.F. and B.J. All authors discussed the results and commented on or edited the manuscript. D.J.K. and B.J. share senior authorship of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 and Supplementary Tables 1-3. (PDF 2953 kb)
Rights and permissions
About this article
Cite this article
Füllgrabe, J., Lynch-Day, M., Heldring, N. et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468–471 (2013). https://doi.org/10.1038/nature12313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12313
This article is cited by
-
Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis
Alzheimer's Research & Therapy (2023)
-
Interplay between lncRNA RP11-367G18.1 variant 2 and YY1 plays a vital role in hypoxia-mediated gene expression and tumorigenesis
Cancer Cell International (2023)
-
Nesfatin-1 suppresses autophagy of chondrocytes in osteoarthritis via remodeling of cytoskeleton and inhibiting RhoA/ROCK signal pathway
Journal of Orthopaedic Surgery and Research (2023)
-
Autophagy: Regulator of cell death
Cell Death & Disease (2023)
-
KAT8 acetylation-controlled lipolysis affects the invasive and migratory potential of colorectal cancer cells
Cell Death & Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.