Abstract
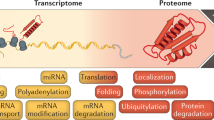
Gene expression differs among individuals and populations and is thought to be a major determinant of phenotypic variation. Although variation and genetic loci responsible for RNA expression levels have been analysed extensively in human populations1,2,3,4,5, our knowledge is limited regarding the differences in human protein abundance and the genetic basis for this difference. Variation in messenger RNA expression is not a perfect surrogate for protein expression because the latter is influenced by an array of post-transcriptional regulatory mechanisms, and, empirically, the correlation between protein and mRNA levels is generally modest6,7. Here we used isobaric tag-based quantitative mass spectrometry to determine relative protein levels of 5,953 genes in lymphoblastoid cell lines from 95 diverse individuals genotyped in the HapMap Project8,9. We found that protein levels are heritable molecular phenotypes that exhibit considerable variation between individuals, populations and sexes. Levels of specific sets of proteins involved in the same biological process covary among individuals, indicating that these processes are tightly regulated at the protein level. We identified cis-pQTLs (protein quantitative trait loci), including variants not detected by previous transcriptome studies. This study demonstrates the feasibility of high-throughput human proteome quantification that, when integrated with DNA variation and transcriptome information, adds a new dimension to the characterization of gene expression regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Stranger, B. E. et al. Population genomics of human gene expression. Nature Genet. 39, 1217–1224 (2007)
Montgomery, S. B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010)
Pickrell, J. K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010)
Stranger, B. E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012)
Kasowski, M. et al. Variation in transcription factor binding among humans. Science 328, 232–235 (2010)
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011)
de Sousa Abreu, R., Penalva, L. O., Marcotte, E. M. & Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 (2009)
Ong, S. E. & Mann, M. Mass spectrometry-based proteomics turns quantitative. Nature Chem. Biol. 1, 252–262 (2005)
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010)
Ow, S. Y. et al. iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355 (2009)
Karp, N. A. et al. Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 (2010)
Li, J., Liu, Y., Kim, T., Min, R. & Zhang, Z. Gene expression variability within and between human populations and implications toward disease susceptibility. PLOS Comput. Biol. 6, e1000910 (2010)
Peng, J., Wang, P., Zhou, N. & Zhu, J. Partial correlation estimation by joint sparse regression models. J. Am. Stat. Assoc. 104, 735–746 (2009)
Lim, J. K. et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 5, e1000321 (2009)
Bonnevie-Nielsen, V. et al. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am. J. Hum. Genet. 76, 623–633 (2005)
Agam, G. et al. Knockout mice in understanding the mechanism of action of lithium. Biochem. Soc. Trans. 37, 1121–1125 (2009)
Sjøholt, G. et al. Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder. Mol. Psychiatry 9, 621–629 (2004)
Pai, A. A. et al. The contribution of RNA decay quantitative trait Loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 8, e1003000 (2012)
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012)
Kersey, P. J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004)
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Frazer, K. A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007)
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003)
Acknowledgements
We thank S. Montgomery for providing gene-level RNA sequencing measurements of CEU LCLs. This work was supported by NIH grants to M.S. and H.T.
Author information
Authors and Affiliations
Contributions
L.W. performed all the experimental work, mass spectrometry-related data analysis and part of the statistical data analysis. S.I.C. performed most of the statistical data analysis. Y.C. performed the protein covariation analysis. D.X. performed the screening of allele-specific peptides. L.J. helped on instrument maintenance. J.L.-P.-T. contributed text and comments to the manuscript. L.W. and S.I.C. wrote the manuscript and participated in detailed discussion of study design and data analysis at all stages of the study. M.S. and H.T. designed and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Mass spectrometry raw data and searching results have been deposited in Peptide Atlas and are available at http://www.peptideatlas.org/PASS/PASS00230.
Supplementary information
Supplementary Information
This file contains a Supplementary Guide, which includes a list of the Supplementary Tables, Supplementary Methods, Supplementary References and Supplementary Figures 1-11. (PDF 1366 kb)
Supplementary Data
This zipped file contains Supplementary Tables 1-8 (see the Supplementary Information file for details). (ZIP 7397 kb)
Rights and permissions
About this article
Cite this article
Wu, L., Candille, S., Choi, Y. et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82 (2013). https://doi.org/10.1038/nature12223
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12223
This article is cited by
-
Mapping genetic variants for nonsense-mediated mRNA decay regulation across human tissues
Genome Biology (2023)
-
Longitudinal multi-omics study reveals common etiology underlying association between plasma proteome and BMI trajectories in adolescent and young adult twins
BMC Medicine (2023)
-
A phenome-wide approach to identify causal risk factors for deep vein thrombosis
BMC Medical Genomics (2023)
-
T cell receptor therapeutics: immunological targeting of the intracellular cancer proteome
Nature Reviews Drug Discovery (2023)
-
Molecular quantitative trait loci
Nature Reviews Methods Primers (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.