Abstract
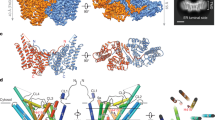
Diacylglycerol kinase catalyses the ATP-dependent phosphorylation of diacylglycerol to phosphatidic acid for use in shuttling water-soluble components to membrane-derived oligosaccharide and lipopolysaccharide in the cell envelope of Gram-negative bacteria1. For half a century, this 121-residue kinase has served as a model for investigating membrane protein enzymology1,2,3,4,5,6, folding7,8, assembly9,10,11,12 and stability1,13. Here we present crystal structures for three functional forms of this unique and paradigmatic kinase, one of which is wild type. These reveal a homo-trimeric enzyme with three transmembrane helices and an amino-terminal amphiphilic helix per monomer. Bound lipid substrate and docked ATP identify the putative active site that is of the composite, shared site type. The crystal structures rationalize extensive biochemical and biophysical data on the enzyme. They are, however, at variance with a published solution NMR model14 in that domain swapping, a key feature of the solution form, is not observed in the crystal structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Van Horn, W. D. & Sanders, C. R. Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu. Rev. Biophys. 41, 81–101 (2012)
Schneider, E. G. & Kennedy, E. P. Phosphorylation of ceramide by diglyceride kinase preparations from Escherichia coli. J. Biol. Chem. 248, 3739–3741 (1973)
Badola, P. & Sanders, C. R. Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J. Biol. Chem. 272, 24176–24182 (1997)
Lau, F. W., Chen, X. & Bowie, J. U. Active sites of diacylglycerol kinase from Escherichia coli are shared between subunits. Biochemistry 38, 5521–5527 (1999)
Pilot, J. D., East, J. M. & Lee, A. G. Effects of bilayer thickness on the activity of diacylglycerol kinase of Escherichia coli. Biochemistry 40, 8188–8195 (2001)
Lahiri, S., Brehs, M., Olschewski, D. & Becker, C. F. Total chemical synthesis of an integral membrane enzyme: diacylglycerol kinase from Escherichia coli. Angew. Chem. Int. Edn Engl. 50, 3988–3992 (2011)
Sanders, C. R. et al. Escherichia coli diacylglycerol kinase is an alpha-helical polytopic membrane protein and can spontaneously insert into preformed lipid vesicles. Biochemistry 35, 8610–8618 (1996)
Gorzelle, B. M. et al. Reconstitutive refolding of diacylglycerol kinase, an integral membrane protein. Biochemistry 38, 16373–16382 (1999)
Vinogradova, O., Badola, P., Czerski, L., Sonnichsen, F. D. & Sanders, C. R. Escherichia coli diacylglycerol kinase: a case study in the application of solution NMR methods to an integral membrane protein. Biophys. J. 72, 2688–2701 (1997)
Nagy, J. K., Lau, F. W., Bowie, J. U. & Sanders, C. R. Mapping the oligomeric interface of diacylglycerol kinase by engineered thiol cross-linking: homologous sites in the transmembrane domain. Biochemistry 39, 4154–4164 (2000)
Wen, J., Chen, X. & Bowie, J. U. Exploring the allowed sequence space of a membrane protein. Nature Struct. Biol. 3, 141–148 (1996)
Smith, R. L., O’Toole, J. F., Maguire, M. E. & Sanders, C. R. Membrane topology of Escherichia coli diacylglycerol kinase. J. Bacteriol. 176, 5459–5465 (1994)
Zhou, Y. & Bowie, J. U. Building a thermostable membrane protein. J. Biol. Chem. 275, 6975–6979 (2000)
Van Horn, W. D. et al. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 324, 1726–1729 (2009)
Lorch, M. et al. How to prepare membrane proteins for solid-state NMR: A case study on the α-helical integral membrane protein diacylglycerol kinase from E. coli. ChemBioChem 6, 1693–1700 (2005)
Caffrey, M., Li, D. & Dukkipati, A. Membrane protein structure determination using crystallography and lipidic mesophases: recent advances and successes. Biochemistry 51, 6266–6288 (2012)
Caffrey, M., Lyons, J., Smyth, T. & Hart, D. J. Monoacylglycerols: The workhorse lipids for crystallizing membrane proteins in mesophases. Curr. Top. Membr. 63, 83–108 (2009)
Li, D., Shah, S. T. A. & Caffrey, M. Host lipid and temperature as important screening variables for crystallizing integral membrane proteins in lipidic mesophases. Trials with diacylglycerol kinase. Cryst. Growth Des http://dx.doi.org/10.1021/cg400254v (2013)
Walsh, J. P., Fahrner, L. & Bell, R. M. sn-1,2-diacylglycerol kinase of Escherichia coli. Diacylglycerol analogues define specificity and mechanism. J. Biol. Chem. 265, 4374–4381 (1990)
Bohnenberger, E. & Sandermann, H., Jr Lipid dependence of diacylglycerol kinase from Escherichia coli. Eur. J. Biochem. 132, 645–650 (1983)
Li, D. & Caffrey, M. Lipid cubic phase as a membrane mimetic for integral membrane protein enzymes. Proc. Natl Acad. Sci. USA 108, 8639–8644 (2011)
Matte, A. & Delbaere, L. T. J. ATP-binding motifs. eLShttp://dx.doi.org/10.1002/9780470015902.a0003050.pub2 (2010)
Krissinel, E. On the relationship between sequence and structure similarities in proteomics. Bioinformatics 23, 717–723 (2007)
Ullrich, S. J., Hellmich, U. A., Ullrich, S. & Glaubitz, C. Interfacial enzyme kinetics of a membrane bound kinase analyzed by real-time MAS-NMR. Nature Chem. Biol. 7, 263–270 (2011)
Walsh, J. P. & Bell, R. M. sn-1,2-diacylglycerol kinase of Escherichia coli. Mixed micellar analysis of the phospholipid cofactor requirement and divalent cation dependence. J. Biol. Chem. 261, 6239–6247 (1986)
Martin, J. W., Yan, A. K., Bailey-Kellogg, C., Zhou, P. & Donald, B. R. A geometric arrangement algorithm for structure determination of symmetric protein homo-oligomers from NOEs and RDCs. J. Comput. Biol. 18, 1507–1523 (2011)
Martin, J. W., Yan, A. K., Bailey-Kellogg, C., Zhou, P. & Donald, B. R. A graphical method for analyzing distance restraints using residual dipolar couplings for structure determination of symmetric protein homo-oligomers. Protein Sci. 20, 970–985 (2011)
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, 370–376 (2012)
Doublié, S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 363, 91–108 (2007)
Caffrey, M. & Porter, C. Crystallizing membrane proteins for structure determination using lipidic mesophases. J. Vis. Exp. 45, e1712 (2010)
Li, D., Boland, C., Walsh, K. & Caffrey, M. Use of a robot for high-throughput crystallization of membrane proteins in lipidic mesophases. J. Vis. Exp. 67, e4000 (2012)
Li, D., Boland, C., Aragao, D., Walsh, K. & Caffrey, M. Harvesting and cryo-cooling crystals of membrane proteins grown in lipidic mesophases for structure determination by macromolecular crystallography. J. Vis. Exp. 67, e4001 (2012)
Fischetti, R. F. et al. Mini-beam collimator enables microcrystallography experiments on standard beamlines. J. Synchrotron Radiat. 16, 217–225 (2009)
Evans, G., Axford, D. & Owen, R. L. The design of macromolecular crystallography diffraction experiments. Acta Crystallogr. D 67, 261–270 (2011)
Cherezov, V. et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 µm size X-ray synchrotron beam. J. R. Soc. Interface 6 (suppl. 5). S587–S597 (2009)
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Stokes-Rees, I. & Sliz, P. Protein structure determination by exhaustive search of Protein Data Bank derived databases. Proc. Natl. Acad. Sci. USA 107, 21476–21481 (2010)
Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D 64, 125–132 (2008)
Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990)
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Painter, J. & Merritt, E. A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Diller, D. J. & Merz, K. M., Jr High throughput docking for library design and library prioritization. Proteins 43, 113–124 (2001)
Acknowledgements
We acknowledge support from Science Foundation Ireland (grants 07/IN.1/B1836 and 12/IA/1255) and the National Institutes of Health (grants GM75915, P50GM073210, U54GM094599) and FP7 (COST CM0902). We thank C. R. Sanders for providing E. coli strain WH1061 and for his collegiality. The assistance and support of beamline scientists at the Advanced Photon Source (23-ID) and Diamond Light Source (I24) are gratefully acknowledged. We thank R. Sanishvili for assistance with zinc analysis, C. Boland and J. Tan for help with diffraction data collection, and A. Coughlan for help with lipid synthesis.
Author information
Authors and Affiliations
Contributions
D.L. produced, purified, crystallized and functionally characterized the protein and its variants, collected and processed diffraction data, refined and analysed the structures, and helped write the manuscript. J.A.L., D.A. and V.E.P. collected and processed data, solved, refined and analysed the structures, and helped write the manuscript. L.V. helped process data, solve and analyse structures and write the manuscript. M.A., C.D. and V.E.P. helped with protein and crystal production. C.P.K. performed docking. S.T.A.S. provided 7.8 MAG for crystallization. M.C. was responsible for the overall project strategy and management and oversaw manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Tables 1-2, Supplementary Figures 1-15 and additional references. (PDF 2723 kb)
Rights and permissions
About this article
Cite this article
Li, D., Lyons, J., Pye, V. et al. Crystal structure of the integral membrane diacylglycerol kinase. Nature 497, 521–524 (2013). https://doi.org/10.1038/nature12179
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12179
This article is cited by
-
Structure of membrane diacylglycerol kinase in lipid bilayers
Communications Biology (2021)
-
Global response of diacylglycerol kinase towards substrate binding observed by 2D and 3D MAS NMR
Scientific Reports (2019)
-
The Transmembrane Conformation of the Influenza B Virus M2 Protein in Lipid Bilayers
Scientific Reports (2019)
-
LILBID and nESI: Different Native Mass Spectrometry Techniques as Tools in Structural Biology
Journal of the American Society for Mass Spectrometry (2019)
-
The cubicon method for concentrating membrane proteins in the cubic mesophase
Nature Protocols (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.